I never did high-impact research. I graduated in the wrong stable for that, to get a head start in plant phylogenetics, you need to make at least your Ph.D. with one of the big players. My official Ph.D. supervisor was a palaeobotanist, my Diplom-degree (now replaced by its international equivalent, master of science, but a Diplom was more work and required (much) more time and skill than a master's degree) was in geology-palaeontology (I put my Diplom-thesis on my homepage and it still attracts quite a lot of traffic, despite being written in German). My second and actual Ph.D. supervisor was well-known in her field but a geneticist (working with ribosomal DNA, one realises that most phylogeneticists have no clue about genetics; having a clue gives you a unique perspective on what a phylogenetic tree can possibly show).
Looking back – cross-disciplinary research is fun but doesn't pay off
My Ph.D. thesis (read in full by at least two people) was on "The Mode and Speed of Intrageneric Evolution – A phylogenetic case study on genus Acer L. (Aceraceae) and genus Fagus L. (Fagaceae) using fossil, morphological, and molecular data" and, as required for German Ph.D.'s, is publicly available (159 pp)
 |
| One of the many colour figures in my Ph.D. |
We call it Publikationspflicht. Mine was one of the first at the University in Tübingen simply stored under a stable URN. Before, you would have to print 1000 or 2000 copies for distribution to university libraries at your own expense (one copy goes always to the federal library).
The upside of this was that I wasn't brainwashed by the Holy Church of Cladistics like many of my fellow plant phylogeneticists at the time. And it let me to a dead-end but very interesting career doing actual cross-disciplinary, and cross-organismal, research.
 |
| From Denk & Grimm (Rev. Palaeobot. Palynol., 2009), one of our classic papers. For more background, see The challenging and puzzling ordinary beech – a (hi)story |
In Tübingen, where I started, the lab (now dismantled) was established to study the small ribosomal subunit of foraminifers, so I may well be the only plant phylogeneticist, who has some foraminifer papers in his portfolio. All of which still worth a read.
- Grimm GW, Stögerer K, Ertan KT, Kitazato H, Hemleben V, Hemleben C. 2007. Diversity of rDNA in Chilostomella: molecular differentiation patterns and putative hermit types. Marine Micropaleontology 62:75–90. — the "hermit types" are fun.
- Aurahs R, Göker M, Grimm GW, Hemleben V, Hemleben C, Schiebel R, Kučera M. 2009. Using the multiple analysis approach to reconstruct phylogenetic relationships among planktonic Foraminifera from highly divergent and length-polymorphic SSU rDNA sequences. Bioinformatics and Biology Insights 3:155-177 [open access]. — A paper like no other, I guess, and much underused blueprint (also unique combination: the first three authors include a biologist who made his Ph.D. in foraminiferology, a mycologist with an unusual mathematical knowledge, and a geneticist-geologist).
- Aurahs R, Grimm GW, Hemleben V, Hemleben C, Kučera M. 2009. Geographical distribution of cryptic genetic types in the planktonic foraminifer Globigerinoides ruber. Molecular Ecology 18:1692–1706. — A timely paper, blueprint for others to follow; the journal wanted to extend its portfolio into new organismal groups (still remember, the eagerness of the editor to have this published despite a certain review reluctance ... and it did attract the amount of citations he hoped for).
- Tsuchiya M, Grimm GW, Heinz P, Stögerer K, Ertan KT, Collen J, Brüchert V, Hemleben C, Hemleben V, Kitazato H. 2009. Ribosomal DNA shows extremely low genetic divergence in a world-wide distributed, but disjunct and highly adapted marine protozoan (Virgulinella fragilis, Foraminiferida). Marine Micropaleontology 70:8–19. — Heavily niched, disjunct, land-locked but still no genetic differentiation.
- Göker M, Grimm GW, Auch AF, Aurahs R, Kučera M. 2010. A clustering optimization strategy for molecular taxonomy and its application to planktonic foraminifera SSU rDNA. Evolutionary Bioinformatics 6:97–112. — A fun paper, could have become a standard but molecular foraminiferology was (still is) a small science claim, owned by others.
- Weiner AKM, Weinkauf MFG, Kurasawa A, Darling KF, Kučera M, Grimm GW. 2014. Biogeography of cryptic species in a marine plankton lineage shaped by niche incumbency and historical contingency. PLoS ONE 9:e92148 [open access]. — Didn't want to do foraminifers anymore after I left Tübingen for Stockholm but the Ph.D. student (first author) needed a helping hand.
We published our first paper together in 2002, in a (that time) well-edited and groomed but low impact journal: Plant Systematics and Evolution.
Denk T, Grimm G, Stögerer K [our late technician, she had magic hands when handling E. coli and DNA], Langer M [a foraminiferologist and my predecessor, became professor in Bonn], Hemleben V [my Ph.D. supervisor and boss, retired]. 2002. The evolutionary history of Fagus in western Eurasia: Evidence from genes, morphology and the fossil record. Pl. Syst. Evol. 232:213-236.
Over the time, it became one of the most cited papers in that journal of the 21st century. And still attracts the one or other citation.
But compared to the big players in plant phylogeny, our "impact" sucked. But we gave a shit, we wanted to do interesting things, others shied back from because there was "no impact" in it. And with time...
GoogleScholar
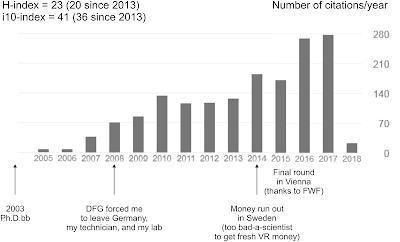
In Stockholm, we lacked soon (official) access to the Web of Science, the public-paid but private-owned citation service, so I looked for alternatives, found GoogleScholar, and got me my own profile to collect my (being cross-disciplinary, it was technically always "our") research to indulge in the meaninglessness of my scientific impact.With the years my status changed from completely irrelevant, which, in professional science, means not cited by others, to not doing so bad (see above, too late to make a career on my own, though; and I was too spoiled by doing what I want how I want to creep up some famous bottoms to give my career a push).
Now that I'm out of business for 1255 days (there's a counter on my homepage), stopped publishing and perished, my profile still looks nice compared with that of career systematic botanists of my age, similar provenance and that got professorships in German-speaking countries. Which is not necessarily a bad thing, because these scores say very little about the research quality of a scientist. It all depends on the circumstances:
- If you have a professorship (chair), you have minions that have to put you on every paper they write; it also makes you interesting for collaboration offers. Some bosses are instrumental in publishing good research, others don't even bother to read it.
- If you are at the right time at the right place, you can corner a market, forcing everyone to work with you or you destroy their papers during "confidential" peer review.
 |
| Single-blind peer review practised by most journals (The review process should be transparent not confidential). |
So, the number of citations can be severely inflated by the number of connections and the stand of a scientist.
GoogleScholar (or the pay-version: Web of Science) records not only the citations of your papers, it also gives a "H-index" and something they call the "i10-index".
The H-index is named after a medicine researcher with the German name Hirsch (Eng.: stag), who noticed that just counting citations is a poor measure. He came up with this alternative.
An H-index of 1 means, you have at least one paper that attracted at least one citation.My H-index now cracked the 30, I'm officially a hard ass (still dead like a Dodo). Of the 50–60 papers, I co-authored, 30 have been cited 30-times or more.
An H-index of 2, two papers, both of which attracted at least two citations.
In GoogleScholar, this includes self-citations, and in the beginning, we were often the only ones citing our papers. Using my GoogleScholar profiles (and forcing my colleagues to get ones, too) it was easy to check which of the papers would need another citation to get the H-index one up.
One of my colleagues in Stockholm (who probably would still be embarrassed me pointing out that he is, and deserves it, among the top-10 of the NRM) found it so nice that he made GoogleScholar profiles for former heads of the Paleobotan at the NRM: Alfred Gabriel Nathorst (current h-index: 23) and Anna Britta Lundblad (current h-index: 9). Our boss (Wikipedia) at the time would be so up in the list that she wouldn't allow us to get her a profile. Danish understatement.
I, on the other hand, took it as a creative exercise, sneaking in a reference that could need another citation to push the stag a bit further.
The H-index and the personal impact factor (PIF) – the number of citations divided by the number of papers you published within a given time period, e.g. the last three years – nowadays cleaned for self-citations (apparently I was not the only one indulging in the sport of citing with purpose) can decide whether your grant application is funded and how you get ranked for a job (in case that job is not already promised to somebody else).
Which brings us to the "i10-index".
Crack the 10 by all means necessary
The i10-index seems to be a very arbitrary measure: it's just the number of papers cited 10 or more times.That may strike one as a low threshold, but most papers published even in the most shiny of science journals such as Science and Nature, never come close to this threshold. Years ago there was a study pointing out that only 25% of their papers generate 90% of these journals' impact. Why it is so hard to get into them unless you have a long track record of being cited by many or come from a booming science market (these days: China, the new number 1 of science).
I only accidently ended up as an co-author of a Nature paper (check out the obligatory Author Contribution statement), and it is (to my personal dismay: What I was not allow to show #1: A neighbour-net of seed plants) my best-scoring paper. Pleased or not, it did increase my general impact.
When still being a professional scientist, I kept a close eye on my GoogleScholar profile numbers, and noticed:
- The more prominent your co-authors in the field of the paper (GoogleScholar links their profiles to your profile), the more citations — that's trivial but also unsettling: Name and fame still trumps content in science, although it is highly fragmented.
- Open access pays off. Especially for those like me publishing in the fringy sciences – intrageneric evolution, palaeobotany, finally systematic (palaeo)palynology – where even the "best" (regarding their impact factor) journals hardly have a JIF (journal impact factor, recorded by another public-financed, private-owned service) > 2.
- When you crack the 10, citations will start to tumble in by themselves from people outside your group and no matter where you published the paper. Copying & pasting citations from other papers is an industry standard. The more a paper pops up in reference lists, the higher the chance it'll be copied & pasted into another one.
You cite your own paper relentlessly to break the 10 citations barrier. There's little risk in sneaking in a citation; reviewers only scout for references you missed (usually papers they published) not the excess ones. In applications, those citations may be vain being self-citations, but it puts your paper on the screen, which may directly give you the desperately needed (or not needed anymore as in my case) x-th citation to push the stag one point higher!And in general:
Advertise shamelessly the papers that still are below 10 or could need another citation to increase your H-index. Point your colleagues to them. Why not trade citations: you cite mine, I cite yours? When reviewing a paper, look out for opportunities to have them cited (PS This is how some of the big shots got their numbers through the roof). Go social media: a paper with four tweets and blog posts already may end up in the top-half of Altmetric scores. And "stakeholders" like such easy-to-access scores.
And when you are on the inclining slope taking off: Consider dropping a co-authorship, you don't need, if the paper cites of a lot of your papers. Self-citations are good for the GoogleScholar profile but those people offering you a job may have access to Web of Science, and there it is just a click to "correct" the H-index and PIF for self-citations. And it doesn't look good when the GoogleScholar values break down to half (or worse). At some point in a scientist's career, being cited by others is more important than being one of many middle authors.PS A last tip for those who wonder whether their profile wouldn't look "good enough" to be shared with the world. You can keep the GoogleScholar profile confidential.


No comments:
Post a Comment
Enter your comment ...