In most plants, the plastome is passed along via the oocyte from the mother, hence dispersed exclusively via seeds (which contain a plant's embryo developing from the zygote). Trees are organisms that play the long card: they take years, some decades, to mature and then they seed their surroundings every year with their offspring. Some of these seeds are minuscule and/or have wings for being carried off by the wind, others are dispersed by migratory birds, but not a few stay in vicinity of the mother plant. As consequence some researchers realised long ago – and many others ignore to the very day – that plastid data finely captures biogeographic trajectories but can be ignorant of species boundaries and poorly sorted: while a wide-spread species can show a plethora of plastid haplotypes, a number of them will be shared with geographically restricted sibling species (or even distant relatives). The plastid signature is decoupled from taxonomy and systematics. If we have nuclear data for comparison, we even can go a step further: from phylogeny.
One of the first, and most cited cases, was Nothofagus (s.str.), the 'false beech'
Acosta MC, Premoli AC. 2010. Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). Molecular Phylogenetics and Evolution 54:235–242.
Premoli AC, Mathiasen P, Acosta MC, Ramos VA. 2012. Phylogeographically concordant chloroplast DNA divergence in sympatric Nothofagus s.s. How deep can it be? New Phytologist 193:261–275.
Acosta and Premoli found that while we have no problems in distinguishing the four of the five modern species of Nothofagus (s.str.) morphologically and using the ITS, the non-coding but transcribed spacers between the nuclear-encoded ribosomal RNA genes, the species share geographically strongly constrained plastid signatures: a northbound, central and southbound plastome.
 |
| Fig. 2 from Acosta & Premoli (2010). Species are abbreviated by three letters. Note the genetic coherence of species when using nuclear data (ITS) and their complete dissolution when using plastid data (covering three intergenic spacers: psbB-psbH, trnL-trnF, trnH-psbA). In the follow up study, the authors confirmed these results using further data. PS This probably also applies to non-South American species, but they have never been studied with such care (last new data produced 2005, Knapp et al. 2005, PLoS Biology; one representative per species) |
A distant, northern hemispheric and species-rich (400+ spp.) relative of the false beeches are oaks. Oaks are notorious in having poorly sorted plastomes telling interesting biogeographic stories when one can rely on extensive sampling (Simeone et al. 2016; Pham et al. 2017; Vitelli et al. 2017; Yan et al. 2019).
 |
| TrnH-psbA-based haplotype network from Simeone et al. (PeerJ, 2018, open access) covering multiple accession from all species of Quercus section Cerris in western Eurasia (white bubbles: East Asian species, data harvested from gene banks). |
Nevertheless, Chinese researchers sequence and publish one hardly interesting plastome after the other claiming its potential to better resolve phylogenetic relationships in the genus or finding a fully resolved species tree (e.g. Yang et al. 2016; Liu et al. 2018; see also Yang et al. 2017; When dating is futile...). Which is impossible because their plastomes don't sort during speciation (Why you should never do a single-species plastid analysis of oaks). Which is obvious once one sequences a second plastome per species from a different provenance (Zhang et al. 2020)
Here, we'll look at my first love, maples (Acer), with respect to a complete plastome phylogeny paper fresh from the press.
A long time ago (for molecular phylogenetics)
In 2006 (Grimm et al., Evol. Bioinform. 2: 7–22, open access), we published an ITS phylogeny of maples using 606 accessions covering all sections and series accepted at the time. In 2008 (Renner et al., Syst. Biol. 57: 795–808, open access), we managed to publish a plastid genealogy – mainly to date it and study inter-continental dispersal – in the flagship journal of phylogenetics: Systematic Biology (the very same journal that rejected our 2006 maple paper after 2nd round of revision; a sort of payback). We didn't combine the plastid data with our earlier nuclear ITS data (Grimm et al. 2006) covering all major lineages and exemplary inter- and intra-species variation because we quickly realised (Renner et al. 2007, Evolution 61: 2701–2719, open access), the plastid and nuclear genealogies are fairly incongruent beyond the tip-groups (now considered sections), which would lead to inflated branch length detrimental for molecular dating.Our studies, like any other phylogenetic study on maples, relied mostly one arboretum samples, which is suboptimal because nature is wild. But we had to work with what we had, not what we wanted. Also, we were just interested in the most general relationships and patterns, and all species of maples are restricted to a specific geographic region and certain habitats: species of maples are pretty well niched. Which is reflected by phenotypes (morphologies). Outside China, species of maples, even from the same section or series, are straightforward to identify will rather little training (oaks are much more variable). Inside China, it can be very difficult because many species hardly differ from each other, morphologically (see description Flora of China) and genetically (just download gene bank data of sects Macrantha, Palmata and Trifida). Species described after 1950 furthermore run the risk of being politically motivated: many common, widespread Chinese species occur also in Japan and have either been described by European, or worse, Japanese "imperialists". Chinese taxonomy tends to split, while North American lumps: some subspecies of the only North American species of sect. Acer, A. saccharum (the 'sugary maple', the actual sugar maple is called A. saccharinum and not related) are not only morphologically but also genetically distinct (Grimm et al. 2007, Plant Syst. Evol. 267: 215–253; yes, 38 pp, quite a lot, rare size for a lowly phylogenetic analysis).
 |
| The final doodle of Grimm et al. (2007, fig. 13) visualising the evolution of Acer section Acer (check out the paper to see what's behind this doodle). |
Age of Big Data
In May this year, over a decade later, Wang et al. (2020, Plant Syst. Evol. 306, advanced online access, paywalled) published the first notable tree based on complete maple plastomes, ...
It is noteworthy that in contrast to the unresolved backbone nodes of phylogenetic trees derived from a few DNA fragments (e.g., psbA-trnH, psbM-trnD, trnL-trnF, ITS) in previous studies (Grimm et al. 2007; Han et al. 2016; Li et al. 2006; Li 2011; Lin et al. 2019; Renner et al. 2007, 2008; Tian et al. 2002), relationships among the sampled 11 sections of Acer were largely well resolved using whole cp-genome data in this study (Fig. 6).
"Few DNA fragments" is a common pun in (Chinese) complete plastome papers to argue why they sequenced the complete plastomes. "Unresolved backbone nodes" is semantically wrong: we established, and discussed, branch support.
It also misrepresents a bit our 2008 plastid tree: only the backbone, the deepest splits had low support (BS < 70, but see graph above), but the major clades' roots (now all recognised as sections) and most sisterclade relationships had moderate (BS > 70) to high (BS > 90) support.
And while we only could access few DNA fragments – the total alignment length in 2008 was 6674 nucleotides, i.e. roughly 5% of the complete plastome – our tree included 66 OTUs (tips) covering all sections and series and a total of 59 accepted species from North America, western Eurasia and East Asia, while Wang et al.'s boosts 28 OTUs ("species"), exclusively from China (including Taiwan, of course, it is a Chinese province from their perspective).
Let's compare the topology of both trees by mapping Wang et al.'s set on our 2008 tree.
A pretty match. There are only two differences.
- Ginnala moved a node down. This may be indeed a better placement, or a sampling phenomenon. Acer tataricum is a wide-spread Eurasian species (or species aggregate, noting the number of accepted subspecies), and the Chinese populations are on the fringe of its distribution range.
- Wang et al.'s complete plastome data struggle with what now is sect. Oblonga (part of the Pentaphylla-Trifida clade), either the complete A. buergerianum plastome is from a inter-sectional cultivation hybrid or simply a misdetermination. Sect. Oblonga includes species of de Jong's earlier ser. Trifida in sect. Pentaphylla often confused with Chinese species of sect. Palmata. Accordingly, Wang et al.'s A. sino-oblongum, according to Flora of China, a species of sect. Palmata despite the name, is resolved within what we called the Pentaphylla-Trifida clade (cf. Grimm et al. 2006).
- "The sect. Negundo diverged firstly..." — Acer negundo represents the first-diverged Chinese species, as seen in our 2008 tree. But, the first diverging plastid clades in 2008 were Clade H, an oddity joining two distantly related (based on morphology and nuclear data) North American and western Eurasian species, and a North American clade with two Japanese species nested inside (Clade G). This is hardly surprising, the oldest known fossils of maples, which can be strikingly similar to the Himalayan Acer caesium or its western Eurasian siblings such as A. pseudoplatanus, are from the Arctic (Arctic Canada, Greenland; something Li et al.'s 2019 trivial out-of-Asia reconstruction also fairly ignores).
- "...followed by sect. Acer..." — refers only to the most ancient member of sect. Acer (see Grimm et al. 2006, 2007). The intriguing bit is lost in the complete plastome tree: most other members of sect. Acer (forming a high-supported 2008 clade, confirmed by Li et al.'s 2019 500-gene data) are deeply nested and one species, A. pseudoplatanus, defies any rule being one of the two species of the first-diverged 2008-clade H. One simply cannot reduce a complex group (see Grimm et al. 2007, cited but not read by Wang et al. publishing in the same journal) like section Acer with a pan-hemispheric distribution and a fossil record possibly going back to the dawn of the genus (e.g. Wolfe & Tanai 1987) to a single accession of a relict species, and then claim to have investigated "phylogenetic relationships".
Ignoring both our 2006 (not cited), 2008 and Li et al.'s (2019) tree (both cited), Wang et al. conclude:
These results suggest that by taking the advantage of next-generation sequencing [...] cp phylogenomics could be used to tackle the tough problem in Acer phylogeny.
They don't. They only show that it is pointless to use NGS resources to exclusively sequence Chinese (micro-)species, while ignoring their counterparts (same species, related subspecies, sibling species) in the rest of East Asia, and – paramount in the case of maples – the much fewer, regarding the number of species, but genetically more distinct (and possibly diverse) species in North America and western Eurasia.Nationalism is generally a bad thing, when it comes to science and phylogenetics, it's a dead-end.
Read carefully, and then look out for interesting bits
Problem-unaware accumulation of plastomes is just a waste of resources. If Wang et al. would have reached out* to get material from critical non-Chinese species, the same amount of plastomes and set of analyses could have been the basis for a very good paper. Identifying those species would have been straightforward from our 2008 tree.
*Would have been allowed to reach out. The People's Republic of China is an authoritarian state ruled by a pseudo-communist party that implemented a highly competitive capitalist system. Science in China always served certain propagandistic needs, in the 60s this meant botanists invented endemic Chinese species by claiming Chinese populations were substantially different from those in neighbouring countries (especially Japan). As obvious from certain papers published by China-based research groups in the last years (especially in Chinese-edited/-reviewed journals), it increasingly seems to serve a nationalist, China-first or China-only, agenda (including botanical papers using no material outside mainland China showing maps with the famous 9-dash line in the South China Sea). I would not be surprised if financial support from the Chinese government means these days that one has to study "Chinese" species.
When the question would be early plastome evolution and biogeographic of the genus, one just would pick one representative (the shortest branched when possible) per well-supported clade and make sure to have all those odd ones outside the trivial clades. If there are any free slots, add species covering missing provenances. Such a sample could have challenged the data-wise trivial out-of-Asia hypothesis put forward by Li et al. (2019): any well-sampled nuclear tree will result in an out-of-China scenario because most species are from China and they are genetically distinct — scattered. This is unsurprising because the modern-day climate in south-western China is most beneficial for relict taxa from greener time periods: especially the mountain forest of Yunnan harbour many plants once widespread across the Northern Hemisphere but today extinct everywhere else, and this likely applies also to the one or other now exclusively East Asian section (intrageneric lineage) of Acer. Why we never did any ancestral area analysis using our ITS data, in contrast to Li et al. (2019), we knew the result would be trivial (China/East Asia) and meaningless. Nuclear data produce nice phylogenies and species trees, resolve inter- and intra-sectional relationships but will tell us little about the biogeographic history of the genus. Plastomes, on the other hand, can carry ancient radiation signatures otherwise lost as documented e.g. in the case of Nothofagus (Premoli et al. 2012) and oaks (Simeone et al. 2016).
 |
| A synopsis of ITS differentiation, the longer the tip, the more distinct the ITS of this group, and the ground-breaking work by Wolfe & Tanai (1987) on the fossil record of Acer (picture from my 2003 Ph.D. thesis). It's funny how well this first doodle still fits data accumulated in the following 20 years. The fossil record of Acer would be in dire need of revision on the background of de Jong's (2002), nuclear data confirmed classification. |
When the question would be how the recently described, genetically intriguing Chinese microspecies A. yangbiense fits in, described as member of sect. East Asian section Lithocarpa but genetically resolved as member of the distantly related circumarctic sect. Acer and assess the difference in the plastomes of species of the same evolutionary lineage, you have to just sample the phylogenetic neighbourhood. In Wang et al.'s case, this would have been sections Acer, Lithocarpus and Marcophylla based on our 2006–2008 results and the recent study by Li et al. (2019) who included A. yangbiense, challenging the original taxonomic treatment. Given that Nepal also has a communist-lead government (currently), it should be possible to establish cooperation and obtain material from the prototypical Acer caesium in addition to the Chinese populations (Himalayan individuals are also found in several arboreta across the world, hence, included in our data set). It's very easy to get material from the European species (most of which are not protected), the biodiversity centre of section Acer. Turkish, Georgian and Iranian maples forming the geographic bridge between the A. caesium complex and its western siblings (mainly species froming Group A, Grimm et al. 2007) would be most interesting, and not impossible to obtain via local contacts. And the North American outpost, Acer saccharum (or its commom subspecies: A. nigrum) can be found everywhere: nurseries and botanical gardens usually have records about the provenance of the seeds cultivated in their arboreta. The North American Acer macrophyllum, the putative sister of the East Asian section Lithocarpa, has not been included in any analysis but would probably be most interesting noting its position in the nuclear trees and its thoroughly primitive morphology (cf. Wolfe & Tanai 1987).
Update 14/7/2020, complete plastomes further confirm our 2008 tree
Another complete plastome phylogeny that "resolves the infrageneric backbone relationships" of maples has just been published in my favourite journal PeerJ.It includes some of the critical species as identified in our earlier papers.
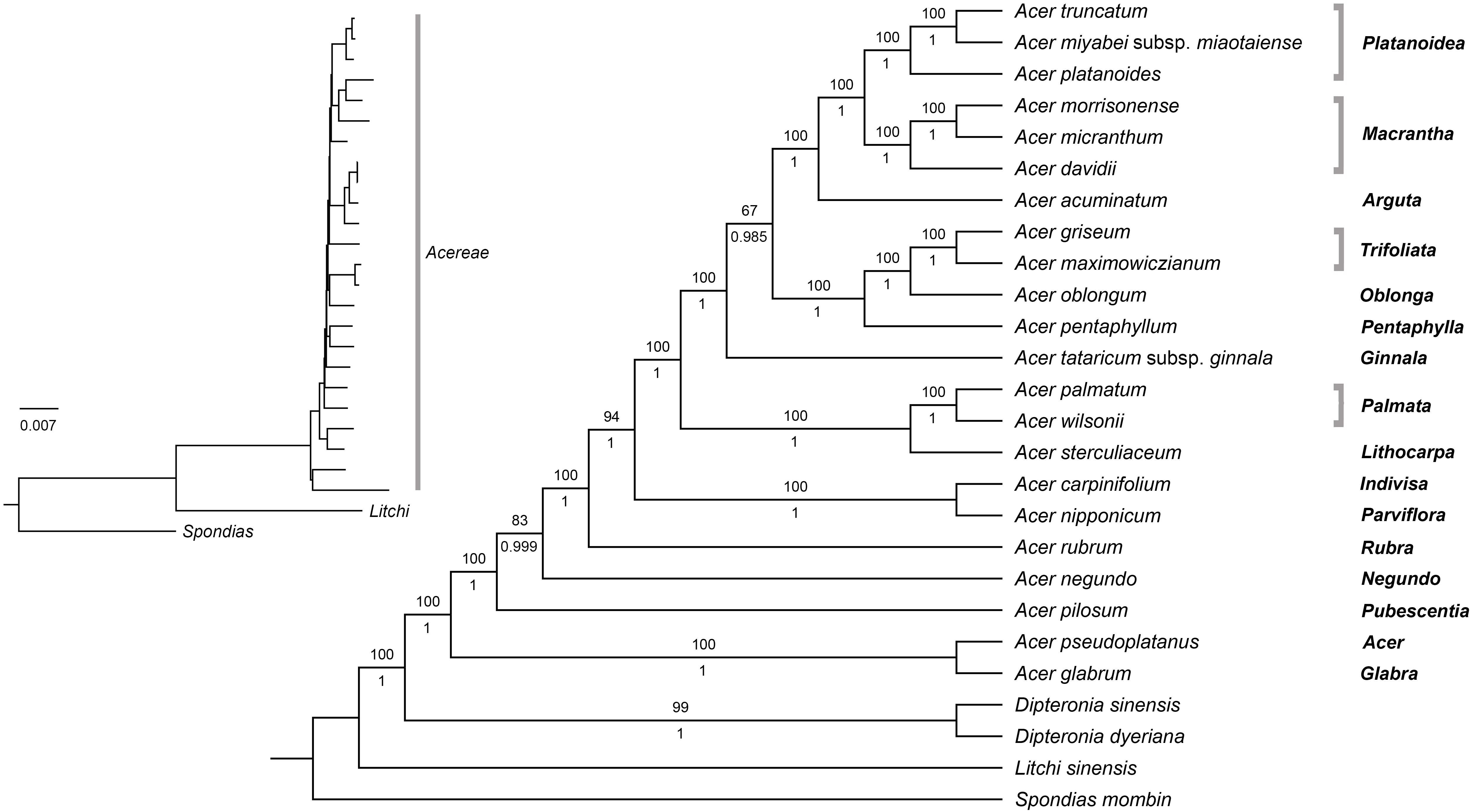 |
| Preferred tree of Areces-Berazain et al. (2020, fig. 7); strikingly similar to our 2007/2008 trees (except for the higher support) |
In contrast to Wang et al. the authors state already in the abstract that:
The plastome-based tree largely supported the topology inferred in previous studies [Renner et al. 2007, 2008; see Discussion] using cp markers while providing resolution to the backbone relationships but was highly incongruous with a recently published nuclear tree [Li et al. 2019] presenting an opportunity for further research to investigate the causes of discordance, and particularly the role of hybridization in the diversification of the genus.
In general, they put more care in their analysis than Wang et al. Unfortunately, Wang et al.'s new plastomes are (could) not (yet be) included in the new paper, so although we find Acer pseudoplatanus, we lack Acer caesium of the most incongruent lineage: section Acer. Complete plastomes are still lacking for any of the other species of this lineage, in which nuclear and plastid genealogies converge. But given the renewed attention, it should be only a matter of time until somebody sequences especially the missing North American species as well, complementing the picture about past distribution patterns.Essential reads on maples
- van Gelderen DM, de Jong PC, Oterdoom HJ. 1994. Maples of the world. Portland, OR: Timber Press. — a book worth buying if you are or want to get interested in maples.
- de Jong PC. 2002. Worldwide maple diversity. In: Wiegrefe SJ, Angus H, Otis D, and Gregorey P, editors. Proceedings of the International Maple Symposion 02. Westonbirt Arboretum and the Royal Agricultural College in Gloucestershire, England: The National Arboretum Westonbirt. p. 2–11. — de Jong's final sectional concept, in line with (produced later!) nuclear data. [PDF]
- Wolfe JA, Tanai T. 1987. Systematics, phylogeny, and distribution of Acer in the Cenozoic of western North America. Journal of the Faculty of Science, Hokkaido University, Series IV: Geology and Mineralogy 22:1–246. — the paper provides the only global analysis of the vast fossil record of Acer; it's a pity the morphological matrix Wolfe & Tanai used for their parsimony analysis has never been published in full and is probably lost to science; the paper only includes the list of scored characters.
References, worth a read
- Pham KK, Hipp AL, Manos PS, Cronn RC. 2017. A time and a place for everything: phylogenetic history and geography as joint predictors of oak plastome phylogeny. Genome DOI:10.1139/gen-2016-0191.
- Simeone MC, Grimm GW, Papini A, Vessella F, Cardoni S, Tordoni E, Piredda R, Franc A, Denk T. 2016. Plastome data reveal multiple geographic origins of Quercus Group Ilex. PeerJ 4:e1897.
- Vitelli M, Vessella F, Cardoni S, Pollegioni P, Denk T, Grimm GW, Simeone MC. 2017. Phylogeographic structuring of plastome diversity in Mediterranean oaks (Quercus Group Ilex, Fagaceae). Tree Genetics and Genomes 13:3 [e-Pub].
- Yan M, Liu R, Li Y, Hipp AL, Deng M, Xiong Y. 2019. Ancient events and climate adaptive capacity shaped distinct chloroplast genetic structure in the oak lineages. BMC Evolutionary Biology 19:202 [e-pub].
- Zhang R-S, Yang J, Hu H-L, Xia R-X, Li Y-P, Su J-F, Li Q, Liu Y-Q, Qin L. 2020. A high level of chloroplast genome sequence variability in the Sawtooth Oak Quercus acutissima. International Journal of Biological Macromolecules 152:340–348.
References, mostly for the trash bin (analysis- and/or text-wise, newly produced data is always good!)
- Li J, Yue J, Shoup S. 2006. Phylogenetics of Acer (Aceroideae, Sapindaceae) based on nucleotide sequences of two chloroplast non-coding regions. Harvard Papers in Botany 11:101–115. — combining what should not be combined, a fine example how to completely mess up a phylogenetic analysis by ignoring every warning flag in the data used, hence, published in the journal of the first author's institution.
- Li J, Stukel M, Bussies P, Skinner K, Lemmon AR, Moriarty Lemmon E, Brown K, Bekmetjev A, Swenson NG. 2019. Maple phylogeny and biogeography inferred from phylogenomic data. Journal of Systematics and Evolution 57:594–606. — nice data but failing to show something really new (as much claimed by the authors), in fact, the tree, "first [to] resolve[d] the basal relationships of Acer" is just a more elaborate version of our 2016 results based on only ITS and not 500 gene regions; the ancestral area analysis (concluding on a Chinese origin) and related discussion is a joke, which may explain why such elaborate data were published in a Chinese journal with politically inflated impact (formely: Acta Phytologica Sinica) and not in a higher impact, more prestigious journal for molecular phylogenies such as BMC Evol. Biol. or New Phytologist. Analysed with a more open mind, the data (no matrix provided as usual) could probably have revealed more (the first author has a long track record of publishing useless Acer phylogenies and obviously no idea what he is dealing with, still).
- Liu X, Chang E-M, Liu J-F, Huang Y-N, Wang Y, Ning Y, Jiang Z-P. 2018. Complete chloroplast genome sequence and phylogenetic analysis of Quercus bawanglingensis Huang, Li et Xing, a vulnerable oak tree in China. Forests 10:587. — a new plastome of a (putative) Chinese microspecies; the paper was obviously not reviewed by someone with the slightest idea about oaks or phylogenetic inference, which is a pity.
- Yang Y, Zhou T, Duan D, Yang J, Feng L, Zhao G. 2016. Comparative analysis of the complete chloroplast genomes of five Quercus species. Frontiers in Plant Science doi:10.3389/fpls.2016.00959. — unlike stated in the title, it completely missed out on the much-needed comparative analysis; again, a pity.
- Yang J, Vázquez L, Chen X, Li H, Zhang H, Liu Z, Zhao G. 2017. Development of chloroplast and nuclear DNA markers for Chinese oaks (Quercus subgenus Quercus) and assessment of their utility as DNA barcodes. Frontiers in Plant Science doi:10.3389/fpls.2017.00816. — in contrast to what the title may imply (and is stated in abstract and conclusions), the provided results and data show the opposite: the complete uselessness of chloroplast DNA markers as DNA barcodes for Chinese oak species. Which, in itself, would have been interesting to elaborate on.







No comments:
Post a Comment
Enter your comment ...