Via a ResearchGate citation alert, I got pointed to a paper by Klak et al. (2020) in PhytoTaxa erecting a new monotypic genus within the Drosanthemeae. I have literally no idea about these plants, but I (obviously in contrast to Klak et al.) now their genetics. A tale of persistent ignorance among renown plant systematicists.
One thing upfront: In contrast to many plant systematicists of my generation (in a wide sense) I crossed during my 15 years as a professional scientist, I don't think the purpose of classification is to rid our systematics of "non-monophyletic" taxa (as done, e.g. by the Angiosperm Phylogeny Group). Not a morphologist myself but a profound admirer of field-seasoned (!) taxonomists, I always believed, we should include molecular evidence to refine the traditional morphology-based concepts, and not to discard them because of some "high-supported" clade in a molecular phylogenetic tree. Which always may be, decreasingly so but still, a data-, method- or model-inflicted branching artefact. Having work with palaeontologists, I also don't see the point in naming molecular clades that have nothing visible in common, hence, are useless for looking back in time.
But with the Molecular Revolution reaching botanical systematics, a generation formed who has still rather a lot (not as much as field-taxonomists) morphological expertise but little idea about the molecular data they are working with and the implicit and explicit assumptions of tree-inference. Hence, rather blindly believe their inferred, single phylogenetic tree. We crossed many of those since we soon straddled from the beaten path; we (more or less) successfully battled them down during the usual "single-blind confidential peer-review".
Publishing peer-review histories as practised by e.g. PeerJ has taken a lot of the vengeful fire out of this battle behind the curtains of scientific publishing. Just check out the peer review history of our papers on Loranthaceae (pissing off Dan Nickrent, nigh-expert on epiphytes, he obviously reviewed our palynological paper on the family, but probably left the deed to review the follow-up published in PeerJ to someone less eminent, and imminent) but also on a until that date neglected but very beautiful southern African genus—Drosanthemum:
- Peer-review history for Eocene Loranthaceae pushes back divergence ages for major splits in the family; see also the related post: Trivial but illogical – reconstructing the biogeographic history of the Loranthaceae (again)
- Peer-review history for Phylogenetic relationships in the southern African genus Drosanthemum (Ruschiodideae, Aizoaceae)
 |
| How can you not be attracted by such flowers? Behold Drosanthemum (Liede-Schumann et al. 2020, fig. 1) |
Certain South African flowering plants are the fief (area of expertise) of Peter Bruyns, and in course of my travels to South Africa chatting with colleagues down there (totally alien people tend to easily open up to me, because I call bullshit, when I see it, no matter the name on it—bad career advise, but very profitable when it comes collecting anecdotes), I came to the understanding that working in his fief leaves you two options:
- You invite His Eminence to be co-author; get the result, He expects, and thy paper is to be published. Few dare (in syst.-bot. circles) to reject a paper co-authored by a Lord of the Realm (my stag has, these days, more ends than his; Nickrent's antlers nearly is as big as our's combined).
- Don't and He'll may be the anonymous reviewer of your paper and do whatever lies in His power to undermine its publication. Especially if you are not in line with the work He condoned with his co-authorship, or as friendly and pleased with the result, reviewer.
I don't blame those people misusing their stand in academic cycles, Gelegenheit macht Diebe: All natural sciences have and some are still a male-dominated world, systematic botany is one of the softests, and the bigger sharks better eat the smaller ones before they become a threat. No-one publishes 20+ research papers a year based on his genuine work. Be nice, and you put a lot of effort in one reviews after the other without being recognised for all your instrumental work and much-needed expertise. Be not nice, and you'll get offered co-authorships.
But like Nickrent's bunch (Trivial but illogical…), also Bruyns people they never look at their sequence data, they only look at the inferred trees, worse, their cladograms. If they don't like the outcome, they may delete tips or genes until it fits their expectations. Not surprisingly, I've never crossed a single phylogenetic paper published or co-authored by Bruyns (or Nickrent or anyone else of that syst-bot Rotary club of elderly white males) that would have impressed me only to the slightest. In fact, all those cladograms fall short of addressing critical issues one has when studying low-level evolution, phylogenetic relationships so very close to the speciation processes: amplitude of signal, its tree-likeness, the a priori discriminative power of the data and each combined genetic marker, the coexistence of primitive and evolved sequence variants, conflicts between nuclear and plastid data, and the many modes of speciation in plants. But the only Bruynsian data I looked at closely is that of the Ruschioideae, to which Drosanthemum belongs, to help out friends in Bayreuth and honour the work of a late botanist who dedicated a good deal of her life to collect and study this genus: Heidrun Hartmann (the English-Wikipedia is a stubby stub, hence, the German). I ended up co-authoring two papers on Drosanthemum, both of which I'm quite satified with.
- Liede-Schumann S, Meve U, Grimm GW. 2019. New species in Drosanthemum (Aizoaceae: Ruschioideae). Bradleya 37:226–239.
- Liede-Schumann S, Grimm GW, Nürk NM, Potts AJ, Meve U, Hartmann HEK. 2020. Phylogenetic relationships in the southern African genus Drosanthemum (Ruschioideae, Aizoaceae). PeerJ 8:e8999.
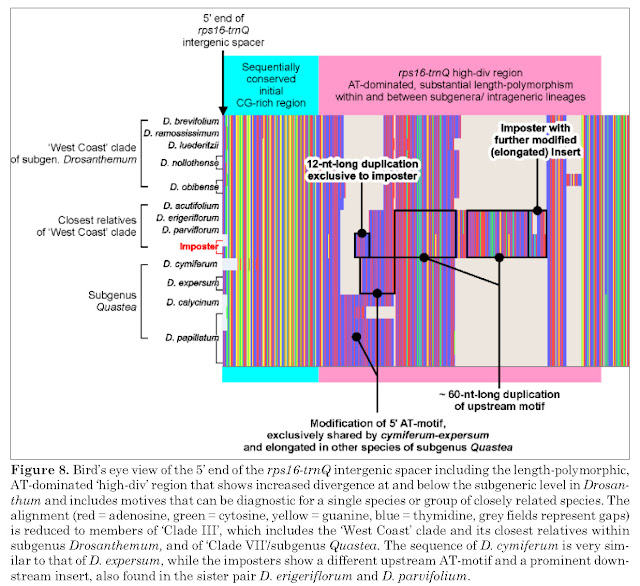 |
| Knowing one's data, from Liede-Schumann et al. (2019): the “imposter” is an individual where the genetype and phenotype doesn't match up, something very rarely observed in Drosanthemum. |
Hence, after having read the abstract, I couldn't help to comment on the paper right away on ResearchGate. Which I'm going to reprint here, but adding figures (naturally) and a bit of lore.
My comment on “previously published facts”
“The results confirm some previously published facts, e.g. that D. zygophylloides is sister to Drosanthemum”—Klak et al. (2020), Abstract.
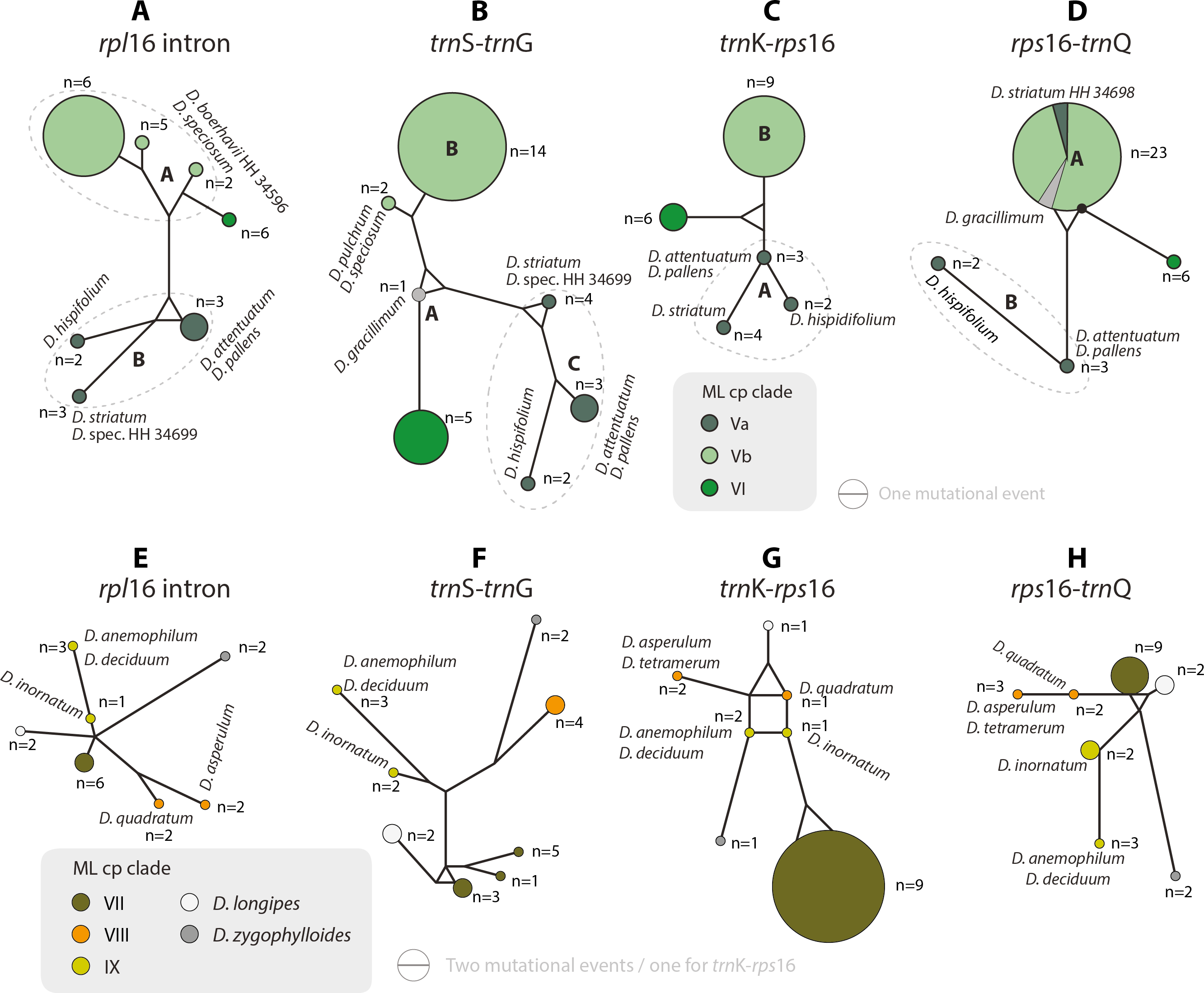
Nice. But, no. We demonstrated — in our paper, cited by Klak et al. and published in the same year — using a combination of tree- and network inferences and in-depth exploratory data analysis that the placement of D. zygophylloides in earlier plastid-based phylogenetic trees by Klak et al. on the subfamily is a branching artefact: ingroup-outgroup long branch attraction. This obviously is still the case.
What we did, and Klak et al. would need to make better
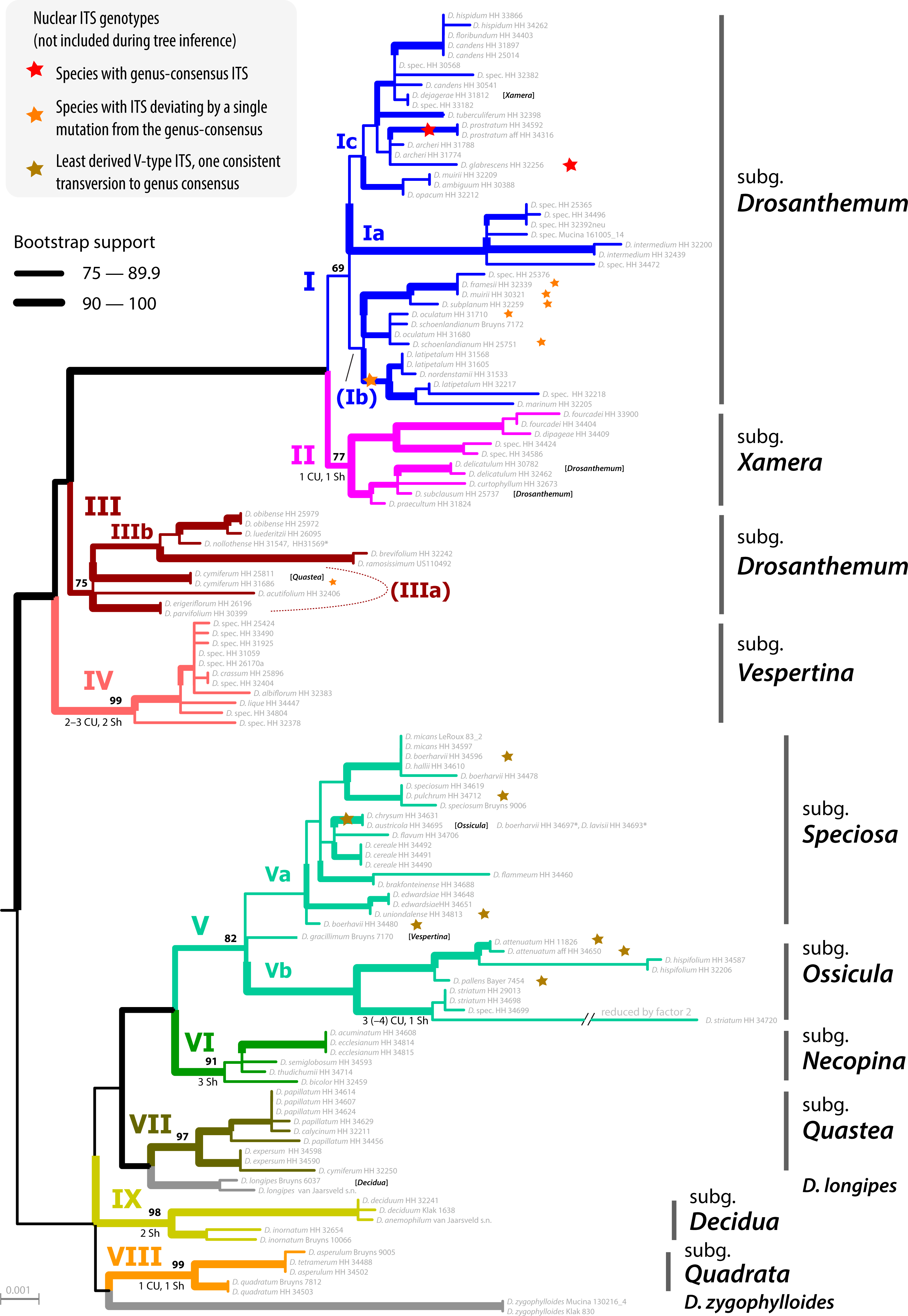
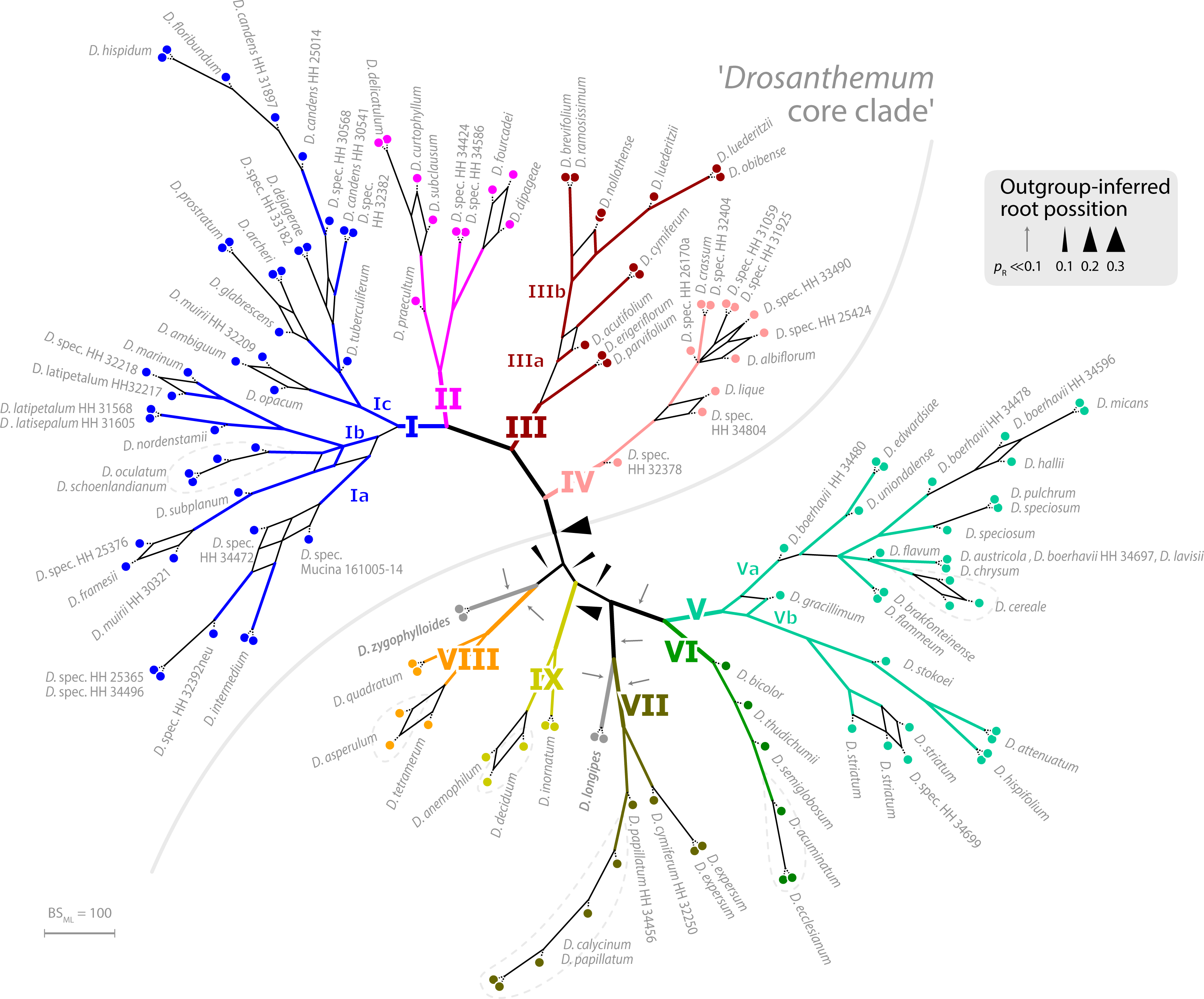
I have no access to paywalled literature (PhytoTaxa is not really a commonly subscribed journal, too nichy, so nichy, you don't even find the paper on Sci-Hub) but I'd be very surprised, if a paper published in PhytoTaxa by very conservative scientist like Klak and alike did what we did to explore the signal in plastid (and nuclear-ITS) data of Drosanthemeae to test their hypotheses. Analyses that, as a whole, are so easily discarded by the authors. Authors, who so far based all their phylogenetic musings on cladograms (often parsimony-based), regarding any "well or high-supported clade" (common threshold: BS support > 70; PP > 0.95) as sufficient and necessary criterion for holophyly, monophyly in a strict sense—inclusive common origin(s).
- maximum likelihood tree inference (Liede-Schumann et al. 2020, fig. 2) and full bootstrap analysis focussing on ambiguous signal by incorporating bootstrap consensus networks (L.-Sch. et al. 2020, fig. 3);
- explicit root testing (main results included in fig. 3) using the evolutionary placement algorithm implemented in RAxML and sistergroup taxa as queries; showing that only the longest branching outgroup taxon, Conophytum calculus (tested were 49 members of the sister clade), was connected to the D. zygophylloides branch (root scenario 3), and finding a low probability (in a mathemetical sense) for this rooting alternative: full results in can be found in L.-Sch. et al. (2020, supplemental information 4);
- in-depth, sequence-based exploration of mutational patterns and plastid differentiation processes, including identification of ancestral and derived (evolved) gene-wise haplotypes (L.-Sch. et al. 2020, figs 4–5);
- cross-checking against nuclear data (ITS region of the 35S rDNA), which cannot resolve interspecies relationships but also lacks any evidence for what Klak et al. call "previously published facts"; instead the ITS of this species can be derived from the rest of the genus' main ITS types, representing a satellite "ribotype" (if you want) of ITS variants diagnostic (evolved within the lineage) for Drosanthemum clades 3 and 4 (L.-Sch. et al. 2020, fig. 6 and supplemental information 3.
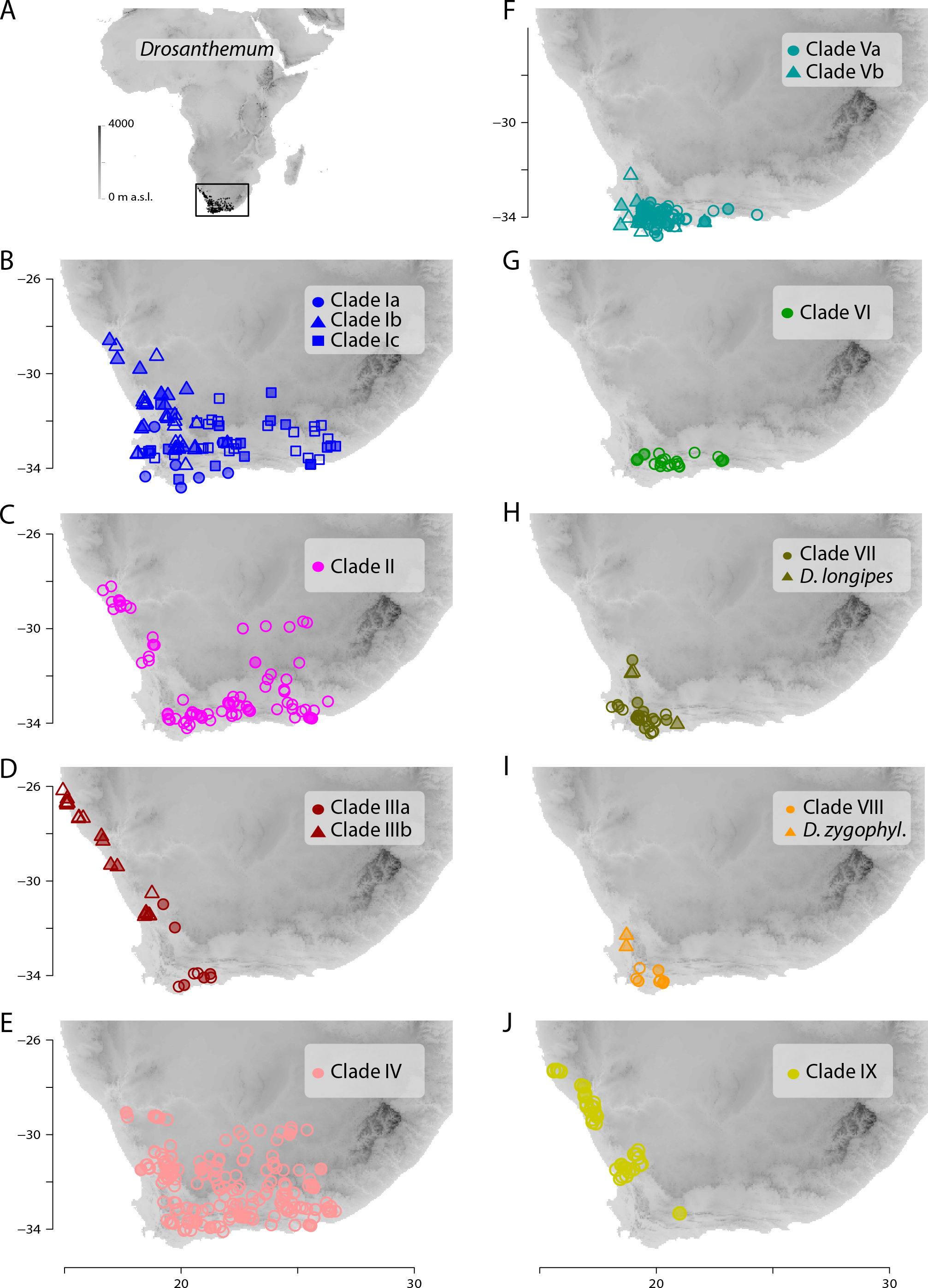 |
| Distribution of the found plastid clades viz subgenera. How likely is it that a monotypic genus survives in the centre of biodiversity of its sister? |
In short, there are multiple lines of molecular evidence showing that while D. zygophylloides is a strongly diverged (evolved), phylogenetically somewhat isolated member of Drosanthemum (possibly closest related to subg. Quadrata), there's no evidence to assume it's an early diverged sister of the entire rest (making subg. Quadrata the first-diverging lineage within Drosanthemum acc. Klak et al. See also the discussion in our paper.
The species evolved from the ancestral species pool of Drosanthemum, specifically the lineage that differentiated into clades V–IX, a morphologically heterogenous group of genetically well-sorted subgenera. It is the product of "budding evolution" not cladogenesis in course of an initial dichotomy. And this is not a “previously published fact” but a well-tested, in-depth analysed biological and evolutionary reality.
But Klak et al. (2020), in their inaccessible paper, may have provided further unrefutable proof. I hope it's more than just another cladogram adding another genes to enfore ingroup-outgroup long-branch attraction. By the way, our data is freely accessible via DataDryad (PeerJ has a strict open data policy, one reason I like the journal), something Bruyns, Klak and coworkers never do: providing the data and (trimmed?!) matrices behind their trees to the public. You don't want to spill the treasures of the Realm to the mere peasants, do you?
What if…
If we entertain Klak et al.'s “published fact” (albeit tested and rejected) that D. zygophylloides is the sister to all other Drosanthemum, i.e. that the (real-world) last common ancestors (LCAs) of the new monotypic genus, Lemonanthemum, and the remaining Drosanthemum (fide Klak et al.) have been mutually exclusive sister species, there must have been some reticulation event. How else would Lemonanthemum picked up the ITS variant of one sublineage within Drosanthemum. Like the ITS types of its close relatives, the species of subg. Quadrata (Clade VIII) its ITS type can be derived from an ITS variant found in a species of Clade III (one of the plastid clades collecting species of the paraphyletic subgenus Drosanthemum; details can be found in L.-Sch. et al. 2020, supplemental information 4 providing a series of statistical parsimony haplotype networks with different tip samples). Which, technically, would render both genera paraphyletic: Drosanthemum s.str. doesn't include all descendants of the last common ancestor, from which all its ITS variants were inherited; Lemonanthemum is the descendant of a lineage that was sister to all other Drosanthemum (reflected in its plastome, when rooted with relatively distant outgroups) but also shares an inclusive common origin with a part of Drosanthemum; the part from which it got its modern-day ITS variant.
Or, more precisely, epi- and periphyletic using Wheeler's (2014, Phyletic groups on networks. Cladistics 40:447–451) categories for reticulation-aware cladistics.
Rooting the ITS differentiation pattern with D. zygophylloides sends further ripples down the entire Drosanthemum s.str. phylogeny: the (near)consensual ITS variants found in species of Clade I (comprising all other members of subg. Drosanthemum) and Clade V (subg. Speciosa and Ossicula) must have evolved in parallel, or "captured" by secondary contact/ inter-subgeneric introgression. Ockham's Razor says, our rooting makes more sense than Klak et al.'s “previously published fact” regarding the minimum necessary number of reticulations.
It would be very easy to proof us wrong: one extracts the plastid mutation patterns exclusively shared by all Drosanthemum except for D. zygophylloides, the new sole species of the novel Lemonanthemum.
If Klak et al.'s assumption is true, there must be genetic synapomorphies to back the hypothesis of mutual holophyly, mutually exclusive common origins, of the now two genera. Especially so, since D. zygophylloides is possibly the genetically most-distinct of all Drosanthemum (hence, it's vulnerability for long-branch attraction inference artefacts): there are plenty of mutations only found in this species and not in any other Drosanthemum. But there's not a single one shared by all those other Drosanthemum, distinguishing their ‘clique’ from all other Ruschioideae including the novel Lemonanthemum. There are conserved, consisten mutational patterns, potential genetic synapomorphies in a cladistic sense, distinguishing between Clade I–IV (subg. Drosanthemum, Verspertina, Xamera) and the rest. Our clades are not just inferences, they are supported by character splits in the data. I know, because we looked at our data and behing the clades in our tree, to extract as much information as possible (Trees informing networks explaining trees).
 |
| Coding complex length-polymorphic sequence motives (L.-Sch. et al. 2020, supplemental information 2, fig. S2B) |
Leave your ivory towers!
I find it very laudable when classic systematic botanists with a good morphological background engage in molecular-backed taxonomy. Without that, the phenotypic side, classificiation is pointless in plants. But one cannot keep ignoring like done by Klak et al., Nickrent et al. and many others, the data reality of molecular differentiation so close to the speciation coalface:
- the signals are not trivial but complex—necessitating exploratory data analysis,
- plant speciation is not a strictly dichotomous process—which we model with standard trees,
- hence, clades in (especially outgroup-biased) trees are not sufficient, maybe not even necessary criteria regarding (semi-)inclusive common origins: holophyly.
If such a thing exist at the genus-level at all, and in a young, still radiating group and it's not all something in-between para- and holophyly.
Beautifully flowering herbs attracting (more or less specific) pollinators may not be as challenging and promiscuitive as wind-pollinated genera (such as oaks and beeches) but systematic botanists should not cling stubbornly to their long-nurtured tree-naïvity. A documented branching artefact is not a “fact”, it's a factual error! And it doesn't matter how often such an error is reproduced and (re-)“published” in systematical-botanical journals controlled – acting as editors and "anonymous" reviewers – by the very same club of people that clings to the old ways, when a single cladogram was all one needed for a new systematical-botanical paper. With amassing phylogenomic data, this is becoming more and more obvious by the minute: to find the &slquo;true tree’, which may more often than not be a species network rather than a species tree, we need to juggle with all the possible trees. A fresh example (flowering herb, but northern hemispheric):
Stubbs RL, Theodoridis S, Mora-Carrera E, Keller B, Yousefi N, Potente G, Léveillé-Bourret É, Celep F, Kochjarová J, Tedoradze G, Eaton DAR, Conti E. 2022. Whole-genome analyses disentangle reticulate evolution of primroses in a biodiversity hotspot. New Phytologist doi:10.1111/nph.18525.
Holophyly, monophyly in a strict sense, the hypothesis a taxon – the handy drawers, we use to categorise and classify Nature's multifariousness and put a name on them – has a ±inclusive common origin (i.e. including Wheeler's epi- and periphyly) expresses itself in many colours, and many, partly conflicting clades. Especially, if we cannot be sure it's all just incomplete lineage sorting (what we model using coalescent trees) but also peppered by evolutionary reticulation, more or less ancient, species that mixed when they formed, and later, when they came into contact with others. Incomplete lineage sorting often comes with polymorphic common ancestors: hybrid or hybrid-ish origins. As little information the ITS contained for resolving inter-species relationships in the Drosanthemeae (notably, it's more signal than found between most genera of the Ruschieae, their sister clade studied by Klak et al. before we turned their eye on Drosanthemum), one thing is already safe to say: The ITS mutation patterns often fit with the major splits observed in the plastid data, a lot of the conflict can be explained by incomplete sorting and the co-existence of primitive and derived copies in the arrays of the LCAs, but in some tips and subtrees, they already tell different stories, capture different aspects of the evolutionary unfolding of this tribe/genus. Why we didn't combine them with our plastid data, in constrast to studies that established certain “previously published facts”.
A final note on erecting new genera, monotypic or other
Personally I have no issue naming known paraphyla – which the new Drosanthemum s.str. would be, with respect to all available molecular data: Lemonanthemum being an evolutionary offshot of Drosanthemum s.str. – would be totally ok, to cherish the species' genetic and morphological uniqueness. But I thought, and was oftmost reminded of it during review, on conferences and in discussions, that it's largely agreed among plant systematicists to only endorse naming of (putatively) holophyletic taxa?
The reason why Klak et al. erected only one new genus, for a species they seem to hold very close to their scientific hearts, and not directly the other subgenera (to ensure holophyly irrespective of the placement of the root), is not that they are harder to grasp, most of them are phenotypically as coherent as they are genetically (see Liede-Schumann et al. 2020). There only one problem, a most unfortunate one (regarding priority and stuff): subgenus Drosanthemum, the morpho-systematic group including the type species of the genus. While genetically coherent, it's morphologically hard to diagnose (as a holophylum, very easy as an explicit paraphylum), possibly showing a primitive (most “plesiomorphic”) phenotype. They simply may have not evolved as much as others of the genus: that Clade I and III have similar morphologies because of genus' symplesiomorphies. Some a their species (coincidence or not?) have an ITS sequence which is identical to the consensus of all Drosanthemeae…
But, with respect to the root-position ambiguity rather than dismissing it (based on one's biased cladogram), they could have suggested to split the genus into seven probably holophyletic genera.
Seven genera would not be so weird: remember, genetically the Drosanthemeae can easily compete with the many genera of the Ruschieae, their sister clade, sequenced by Klak et al. Splitting this genus into seven genera may even better reflect its genetic diversity than keeping those lineages in one, just because their morphology is less diverse. Why they didn't do that? My guess, because that would have required to put some real work – priorities, diagnoses and all – and thinking – acknowledging that it's difficult to infer a root in a fast ancient radiation setting – into the paper, they were busy to publish, without the risk of any real review, in PhytoTaxa.



No comments:
Post a Comment
Enter your comment ...