In part 1, I introduced and dissected two phylogenomic trees of maples, a complete plastome-based (Yu et al. 2022) and a nuclear one (Li et al. 2019). The former could have revealed some interesting evolutionary patterns, if one only would have compared it to the latter. The latter would have made a perfect data set to fix the Acer systematics, including defining thresholds for what to expect from a maple section (→ part 1). But naturally there's more the authors could have found out, if they would have not been so determined to better support what was already known for over a decade. The commonly ignored non-trivial aspects of phylogenomic trees.
What I'd have done with such data at hand
What do you do with 500+ nuclear genes of a genus that has a long-documented non-trivial, possibly reticulate evolutionary history? Fishing for inter-gene incongruence, of course. Just needs an annotated matrix: a partition file saying which of the 421723 bp in Li et al.'s (2019) matrix represent gene #1–#500. But more importantly, I would have mapped the plastomes on my fancy nuclear coalescent/multi-gene tree.
Do you still capture, or are you sorting already?
Why? The plastomes are the genomes inherited from the mothers over relative short distances (discussed in Renner et al. 2008; see also Scientia-ex-machina…). They can tell you where a lineage originated, specifically, where the LCMs, the ‘last common mothers’ stood (see last section of this post for a hypothesis based on other people's phylogenomic trees). Which Li et al. (2019) noticed but ignored when interpreting their results and setting up their analysis.
However, the Northern Hemisphere disjuncts identified by the plastid DNA sequence data differ drastically from nuclear genes and morphology. For example, species of section Parviflora [s.l.] do not form a clade, nor do those of section Spicata [s.l.] or Negundo [s.l.;→ part 1] (Renner et al., 2008). The conflict may have resulted from the fact that the plastid genomes are maternally inherited (Corriveau & Coleman, 1988) and chloroplast capture or incomplete lineage sorting may have happened producing genes trees that are incongruent with species trees.—Li et al., 2019, last page.
You can only capture a plastome that is (was, in the contest of historical biogeography) nearby. If two sister lineages (nuclear-morphology-wise) have to (nigh-)different plastomes, their respective LCA's, their ‘last common ancestors’, cannot have the same geographic origin: to capture a substantially different plastome, I need to first disperse into the area of a distant sister lineage. And incomplete lineage sorting (at this level) implies a polymorphic ancestral plastid gene pool. Which you only have if the LCA was widespread (a multi-area species), and, following a (large-scale) vicariance event, its descendants sorted their plastomes geographically, based on their disparite point-of-origins, and not phylogenetically. Hence, they appear to be incompletely sorted.
Here's the minimum-effort map Li et al. could and should have included:
 |
| Li et al.'s “Ancestral area inferences using statistical dispersal and vicariance analysis (S‐DIVA) and dispersal and extinction cladogenesis (DEC) methods” leading to a non-sensical “Asia [meaning East Asia] might be the most likely ancestral area of Acer as proposed by Wolfe and Tanai.” Ignored by Li et al. (2019): the plastomes of these species (including Li's earlier plastid-based studies on certain subgroups) beg to differ: Annotated to the right are the major plastome lineages as identified by Renner et al. (2008) and traceable in later complete plastome studies (Areces-Berazain et al. 2020; Yu et al. 2022). BFLDD—inferred by Li et al. (2019) but implausible back-and-forth long distance dispersals (for more see Scientia-ex-machina…) |
Whether captured in course of dispersal and secondary contact or incompletely sorted, if two sister lineages have substantially different plastomes, their respective point of origins cannot point to the same area. Because they don't share an exclusive to both LCM—a ‘a last common mother’ tree/ population. The sections characterised by non-palmate leaves and carrying B plastomes – exclusively East Asian sects Arguta and Cissifolia – do share a LCM with the A plastome carrying equally non-palmate sect. Macrantha (originally East Asian; all three part of Wolfe & Tanai's ‘Spicata group’ and the palmate, but strongly different sect. Platanoidea—cross-Eurasian, with a strong preference for higher latitudes in East Asia. Renner et al. (2008; using several dating experiments, results tabulated as supplementary information) estimated a median minimum age of ~33.5 Ma [42–24 Ma] for the MRCA, the hypothetical ‘most-recent common ancestor’, the hypothetical LCM population, of A+B plastomes. Arguta and Cissifolia are part of a larger nuclear clade (= ‘palmatoid cluster’, cf. Grimm et al. 2006; ≈ ‘Spicata group’ acc. Wolfe & Tanai 1987) of East Asian origin according Li et al. (2019). Sect. Palmata (including a single, pretty nested North American species) and the East Asian species of Spicata s.l. (= ser. [sect.] Caudata [→ part 1]) have D plastomes; their plastome MRCA was slightly younger (30 [40–21] Ma). The MRCA of A–D plastome clade was placed ~10 myrs earlier. This would place the initial vicariances of the plastid gene pool of today's most-diverse Acer section-lineages in the (early to mid-)Eocene. A very old picture (basic map done for a talk on my Ph.D. thesis, Grimm 2003; Searching for a research object…).
Plenty of fossils with the fitting phenotypes, distributed across Eurasia, which may include already the LCMs of A to D plastomes. The two (B, D) of four exclusively Eurasian plastomes (A–D) characterising the East Asian members of the ‘palmatoid cluster’ may indeed have their origin in a widespread, cross-Eurasian ancestor, incompletely sorted in the very first phase of maple radiation: A+B may be pointing to a North-East Asian, C to a high-latitude Eurasia/mountains of central Asia (→ Scientia-ex-machina…), and D to a European origin. Or the other way round.
While the plastomes of the North American sister lineages of sects Cissifolia, A. negundo (monotypic sect. Negundo s.str., unique, deep-rooting plastome), and Caudata, A. spicata (monotypic sect. Spicata s.str., E plastome shared with North-East Asian (Japanese) but not related sects Indivisa and Parviflora s.l., diverged earlier. Just by mapping, one can count the captures and incompletely sortings, something an editor and reviewer should have asked for when assessing Li et al.'s paper, given it's focus on biogeographical history (more than a third of the very conscise results, p. 597; 2 of the 5 figures; one of four discussion sections: “4.3 Asia as the ancestral area”).
Disclaimer: You don't want to use the above figure in a publication because one first would need to properly revise the fossil record of Acer, especially the Paleogene one: the group concept pictured here is that of Wolfe & Tanai (1987), which doesn't hold in the light of molecular evidence (as I found out the hard way during my Ph.D., when I tried to reconstruct their morphological character matrix). For instance, sect. Rubra is not part of the lineage leading to sect. Macrantha but related to sects Acer and the Pentaphylla-Trifoliata clade, and a ‘Macrophylla group’ including sect. Acer must be paraphyletic in a rather broad sense. The modern-day Acer macrophyllum, which's leaf type is considered the ancestral prototype for this group, forms the monotypic North American sister lineage of sect. Platanoidea (see Grimm et al. 2006, all confirmed by Li et al. 2009). Mapped on Li et al.'s tree (while citing the paper, they didn't bother about the implications), the ‘Macrophylla group’ would be the very basis from which the ‘Macranthera group’ and ‘Platanoidea group’ evolved. The ‘Spicata group’, however, may be synonymous with the first-diverging modern maple lineage (in Li et al.'s “first fully resolved phylogeny”)—our 2006 ‘palmatoid cluster’.
 |
| A historic doodle summarising my Ph.D. thesis results on maples (Grimm 2003; open access), fossil taxa following Wolfe & Tanai (1987). Note the time scale on the left. |
Riskless dating = pointless dating
Which leads us to another totally unexpected result of Li et al.'s study: “…molecular dating using Bayesian evolutionary analysis by sampling trees (BEAST) indicat[ing] that section diversifications of Acer might have completed largely in the late Eocene and the intercontinental disjunctions of Acer between eastern Asia and eastern North America formed mostly in the Miocene.”
What a waste of “500+ nuclear loci” and the “first resolved Acer phylogeny”! It's historically trivial that the initial diversification took place in the Eocene, the oldest Acer fossils are from the Paleocene/latest Cretaceaous of the High-Arctic, and the genus has no a bad fossil record in the Eocene, sort-of-raining down from the High Arctic into the palaeomountain ranges of both Eurasia and North America (see e.g. old pic above). Boring already from a 2010 perspective, but still novel-enough for a Q1-journal a decade later.
Scientific fun-fact: One typically ends up with Miocene crown group radiations, when we using too few constraints to node-date a tree with too many tips. Usually they are underestimates, some turned out to be much too young.
Anyhow, surely a huge step forward regarding what those unsatifactory poor-resolution earlier studies showed:
The nine North American species of Acer diverged from their nearest relatives at widely different times: eastern American Acer diverged in the Oligocene and Late Miocene; western American species in the Late Eocene [sic!] and Mid Miocene; and the Acer core
clade, including A. saccharum, dates to the Miocene. Recent diversification in North America is strikingly rare compared to diversification in eastern Asia.—Renner et al. (2008), abstract, paper cited in Li et al. (2019) at 16 places.
 |
| Primary radiations in Acer based on their geographically sorted plastid signatures (minimum estimates after Renner et al. 2008; → Scientia-ex-machina…) |
The tricky bit about dating is not getting the estimates, but interpreting them properly by putting them into context!
Interesting would have been to use these new nuclear-based estimates and compare them 1:1 to the earlier ones from the plastomes and try to put up a holistic scenario for the evolution of maples across the Northern Hemisphere.
For instance for the ‘palmatoid cluster’ carrying all those phylogeny-incongruent A, B and D plastomes. Li et al.'s chronogram (Li et al. 2019, fig. 4, reprinted below) gives a mean minimum age of 63 Ma (lower Paleocene, fixed by oldest Dipteronia fossils) for the MRCA of A/B-plastome carrying sections, which, in their nuclear tree, equals the MRCA of the entire genus. The MRCA of the A-plastome carrying lineages is ~10 myrs younger (53 Ma), the B-carrying MRCA a bit older than the A-carrying MRCA (59 Ma). Since the nuclear tree not only conflicts topologically with the plastid tree, but also flips the general sequence, the A/B-carrying MRCA equals again the genus' MRCA. The MRCA of the D-plastome carrying sections (near-exclusively East Asian) is the very next node, hence, only slightly younger (mean minimum age of 61 Ma), because of the very short, possibly underestimating branches making up the deepest parts of the maple phylogeny. [The data may have a signal amplitude issue. The root-tip length relation in Li et al.'s trees is highly problematic: short roots competing with very long terminal tips; their fig. 1; → neighbour-net shown in part 1.]
The primary evolutionary lineages established and diverged before their plastid pools fragmented. Which, if being reasonably valid estimates, eliminates incomplete lineage sorting as explanation for most of the deep nuclear-plastid incongruence. The lineage-FCAs, the ‘first common ancestors’ must have been cuddling together in a relatively, easy to cover area, since their plastomes hadn't started to drift, genetically. The section-FCAs, evolving much later, must have been very promiscuitive bastards, not being picky regarding the pollination of ±related mother trees. Effectively, all deep-diverging plastomes intermixing with A-B-D-dominated clades were captured, and while incomplete lineage sorting can explain the scattering of A and B plastomes across sects Macrantha and Platanoidea vs sects Cissifolia s.str. and Palmata, it cannot explain why the late-diverging sects Caudata (→ part 1; sister of E-carrying N. American sect. Spicata s.str.) and Lithocarpa (part of the Acer crown group including C-carrying Acer+Pentaphylla-Trifoliata and the sections, and various isolated taxa with other plastomes) share the D plastome with sect. Palmata.
When the new take over the old
A proper, better-informed dating could have also used to revisit once more the hypothesis put up by Boulter et al. (1996; cited in Li et al. 2019, but not when discussing their dating results): that the modern diversity originated not earlier than 35 myrs ago, mixing and messing up everything we see before. Which would pretty much explain the confusing plastid sorting patterns. If Boulter et al.'s hypothesis is true, we don't look at an Eocene diversifications when analysing the nucleome, but post-Eocene isolation, homogenisation and sorting processes of lineages that are monophyletic in a pre-Hennigian sense, but not holophyletic in a strict Hennigian one. We get too old estimates for the primary divergences because we rely exclusively on fossils predating the secondary messing-up as age constraints: we use stem lineages to fix crown groups, i.e. invoke a first-level error.
The fossil fruit assignable to Dipteronia was discovered from the Tertiary North American (McClain & Manchester, 2001). The age of the fossil (60 Ma) was used to calibrate the node [meaning MRCA] of Acer and Dipteronia [leading to a “mean estimated time” of 64.2 Ma]. The age of fossil leaves and fruits (56.5Ma) similar to the species in the clade containing Acer negundo, A. henryi, and A. cissifolium [i.e. sect. Negundo s.l.; reference missing] was used for the node of Arguta and Negundo [56.9 Ma in the dated tree].—Li et al. (2019), Material & Methods, p. 596. [Quite a funny phrasing for a paper coauthored exclusively by U.S. Americans.]
In our 2008 paper, because we relied on selected plastid markers producing more balanaced root vs tip lengths and did not use any additional ingroup constraints (one first would need to revise the fossil record to make sure to use the right ones), our Acer MRCA was not nearly as old as the Acer-Dipteronia MRCA (irrespective of the used age constraints), but ~15 myrs younger, while Li et al.'s (2019) set-up effectively constrained the initial Acer radiations to have happened before 56.5 Ma. Thus, they got 10–15 myrs older divergence ages for the early radiations (but generally younger section-stem and -crown ages; which is a red flag for dating bias; → part 3). Getting older divergence ages than in earlier studies is usually a good thing, but how reasonable is a Paleocene-early Eocene primary radiation of modern maples? Do the reject Boulter et al.'s 35 Ma total turnover hypothesis?
Li et al. (2019) give a latest Oligocene (Chattian) age (mean 24.4 Ma) for the MRCA of the North American A. negundo (sect. Negundo s.str.) and its East Asian sister, sect. Cissifolia. The 30 myrs older Negundo-Cissifolia-similar “fossil leaves and fruits” used as ingroup age priors must then be very early stem fossils of the Negundo-Cissifolia lineage (= sect. Negundo s.l.), maybe even represent the Negundo-Cissifolia FCA. Plus, we already knew a decade earlier that the Negundo-Cissfolia geographic continental split (cross-Beringia) evidenced in their plastid signatures is at least double as old! The North American A. negundo plastome belongs to an ancient, early diverged lineage, much afar from Cissifolia's East Asian B plastome shared with their other sibling, the also East Asian sect. Arguta.
Our oldest estimates, place the divergence of the Negundo-unique plastome from the undifferentiated primordial maple plastome pool in the early Eocene (Lutetian), with the upper bound being 56 Ma. That is in the same time-slice when the ‘last common mother’ of all A–D-carrying lineages lived according to Li et al.'s nuclear dating. Our inferred upper bound exactly matches the age of Li et al.'s sects Negundo-Cissifolia “fossil leaves and fruits” from, naturally, North America (it's never Out-of-Asia). A perfect fit, right? These fossils may have carried the same plastome lineage than the modern-day species A. negundo. But then they are not precursors of section Cissifolia, because Cissifolia B-type plastomes inform us that they have the same point of origin than section Arguta, and both are today restricted to East Asia.
Li et al.'s divergence ages – (pre-)Eocene primary radiation, sectional diversification during the Oligocene-Miocene transition – simply don't make sense in a historical context. Unless, reticulation is added to the equation: secondary contact and pre-sectional lineage mixing (cf. Boulter et al. 1996).
We can be pretty sure our 2008 estimates are rather underestimating: we used node dating with only root constraints and no internal ingroup age constraints. But Li et al.'s may be (grossly) overestimating regarding the deep splits because of a first-level error: assigning morphologically similar foliate stem fossils, an ancient North American maple lineage that evolved the Negundo-type plastomes, to a modern-day crown group, a Cissifolia-type species migrating later into North America and hybridising there with a morphologically similiar cousin to evolve what would become the modern A. negundo. Or, the Cissifolia-FCA was actually a North American migrant and an introgressor of proto-Argutae mother trees, picking up their B plastomes. In either case, the North American Negundo-Cissifolia-similar fossils may be used to constrain the stem age of Negundo or the MRCA of Negundo + Cissifolia plastomes, but cannot inform the stem age of the section Negundo s.l. as used by Li et al. (2019). Maybe Boulter et al.'s were totally right (and Wolfe & Tanai pretty wrong) and the plastids give us the early radiations (along montane temperate niches and cross-Arctic), and the modern-day lineages are the product of Oligocene mixing and sorting, when global cooling triggered the high-time of maples and a new phase of global migration. Too speculative? For sure, too complex for a Q1-journal and standard phylogenomics. But wait for the last part of this long-read to make up your mind.
Li et al.'s data and dating (2019) could have provided another mosaic stone, if they only would have linked their nuclear divergences against the reported plastid ones. After all, it's the first explicit molecular dating of nuclear data. We didn't include our ITS data in the 2008 dating because a) incongruent and b) you cannot use ITS data for dating northern hemispheric trees–it behaves much too unclocky. It would have been timely to use a comparative dating, i.e. use age priors that work for both the nuclear and plastid tree, and make a 1:1 comparison.
Break a tooth from the crown, your Phylogenomic Majesty (→ Trivia), your honour could have just re-used our open data (free-to-download @ TreeBase): we covered more than enough of the leaves than you have in your dated tree!
Like always, it's the tiny, easy-to-overlook things that matter
One really novel thing, we can learn from Li et al.'s “first resolved Acer phylogeny” is that the recently described A. yangbiense (Chen et al. 2003) is not a member of sect. Lithocarpa unlike highlighted in Li et al.'s abstract and indicated by its morphology but a close relative of the (genetically quite isolated; Grimm et al. 2007) Himalayan species of sect. Acer (A. caesium, A. giraldii, the latter typically regarded as a subspecies, even in the splitty Flora of China, Xu et al. 2008, but so far there are no proper genetic data to test this assumption). Or in Li et al.'s own words:
Acer yangbiense of section Lithocarpa (Chen et al., 2003) was included [here] in section Acer with the closest relationship with A. caesium Wallich ex Brandis. … In the Bayesian trees, section Acer (including A. caesium and A. yangbiense) formed a strongly supported clade [and were placed as sister to the remainder of sect. Acer, cf. Grimm et al. 2006, they really kept the text short], while ASTRAL analysis grouped A. caesium and A. yangbiense with series Saccharodendron and Monspessullana [two of the three series of sect. Acer, one holophyletic, the other possibly paraphyletic], and together they [meaning the complete sect. Acer incl. all members of the 3rd ser. Acer, paraphyletic (cf. Grimm et al. 2007)] were sister to the clade of sections Pentaphylla and Trifoliata.—Li et al. (2019), Results, p. 597
Have we forgot to update the abstract, haven't we?
…and A. yangbiense may be included in section Lithocarpa…—Li et al. (2019), Abstract, p. 594
Mistypers happen, but given that this is an actual novel result showing how useful phylogenomics can be for taxonomy, such a mistyper should have jumped to the eye of the expert (co-)authors (→ Trivia), or those of the “…two or more anonymous reviewers from anywhere in the world.” (quote from the journal's author instructions). I qualify for “anywhere in the world” but was never asked to review any paper on Acer because I'm not an expert (I'm really not, taxonomically), nonetheless, I noticed this discrepancy directly (and all the other little mistypings found across the manuscript).
Why is this interesting? It's not just a misplaced species (like in the trees of Yu and colleagues, → part 1) but a Lithocarp-ish (?!) long overlooked or recently emerged (only described in 2003) morphotype. It's the first-known, genetically confirmed discrepancy between morphology and (nuclear) genetics in the genus at the section-/series-level! But it doesn't stop there: Acer yangbiense shares not only its nucleome but also its plastomes (as seen later in Wang et al.'s 2020, and Yu et al.'s trees: → F plastomes) with a taxon (A. caesium s.l.) that is sym-/parapatric in its eastern range (A. giraldii) with A. yangbiense. Although their morphologies associate them with too substantially different cousin lineages: according to Li et al.'s own dating analysis, their fig. 3, the nuclear MRCA of sects Acer and Lithocarpa can be placed in the mid-Eocene, at ~45 Ma! The Aceri and Lithocarpae are very distant cousins.
There are three explanations:
- Acer yangbiense is a member of sect. Acer camouflaging as a Lithocarpa species. This would make it a crucial phenotype to explore further the evolution of the genus because such a shared similarity can only be because of retention of primitive traits (‘plesiomorphies’): it's a(nother) living fossil. Like A. caesium is for its section (Grimm et al. 2007) and possibly for the larger lineage, the ‘aceroid cluster’, if not the genus itself. Acer caesium's leaf morphology is conspicously similar to that of the earliest maple leaves traditionally included in the fossil-species A. arcticum (which may also include non-maple fossils, but in contrast to what Wolfe & Tanai 1987 stated, one can find A. arcticum leaves in the same fossil-beds, e.g. in Greenland, together with prototypical maple fruits: samaras). The Lithocarpae belong to Wolfe & Tanai's apparently distinctly morphologically primitiv (‘plesiomorphic’) ‘Macrophylla-group’; before seperating them, de Jong (1994) placed them as series into the now monotypic, exclusively North American sect. Macrophylla. Like other New World monotypic lineages, A. macrophyllum carries around a pre-A–D-diverged plastome.
- Acer yangbiense is a member of the Himalayan lineage of sect. Acer (our 2007 ‘Group A0’) affected (at some point in the past) by inter-section gene flow involving a Lithocarpa species (extinct or retreated today), leading to its deviant phenotype: a notho-species, a species of hybrid origin. If that's the case, and if Li et al.'s 500+ nuclear loci have the capacity to resolve interspecies relationships, one should find some Lithocarpa genes (as, e.g., can be found in western Eurasian beeches, who were introgressed by an Atlantic-North American lineage at some point in the past; Cardoni et al. 2022, using the data of Jiang et al. 2021).
- Acer yangbiense represents and intrograde or introgressor of the Lithocarpa-lineage: e.g. a Lithocarpa-species invading the Himalayan realm of ‘Group A0’ (Grimm et al. 2007, see below), picking up their back then already F-ish plastome, and, because of frequent backcrossing with proto-A. caesium s.l., replacing their original Lithocarpa nucleotype by a (mainly) Acer nucleotype. Again, just a little EDA – exploratory data analysis – would have sufficed.
When Big-Data Präpotenz meets evolution's coalface
Another classic shortcoming of phylogenomic studies is their ignorance of more detailed works, which, if cited, are typically dismissed as irrelevant, or not cited at all. We did high-resolution taxonomic studies on two sections of Acer: section Acer (Grimm et al. 2007) and sect. Platanoidea (Grimm et al. 2014). The former could have informed Li et al. (2019) to take a bit more care, regarding the most fuzzy part of their “first fully resolved Acer phylogeny”. What to look out for in the leaves: if we have deep reticulation syndromes (nucleome-plastome incongruence like in the ‘palmatoid cluster’), phenotypically wrong microspecies like A. yangbiense), we probably also have rather flat reticulation phenomena, worth exploring, or at least, to point out.
Section Acer was part of our 2006 ‘aceroid cluster’, and sister lineage of the Pentaphylla-Trifoliata clade. Li et al. largely confirm our 2006 results within this subtree. But, using 500+ loci, instead 600+ nucleotides, naturally with much higher support.
If we dive into the clades, it's astonishing in which perfect fit the interspecies relationships are in case of the Pentaphylla-Trifoliata clade, where the 2019 phylogenomic and our 2006 ITS study have about the same tip sample. But in sect. Acer, where the ASTRAL-inferred MSC and the ML-inferred combined tree start to disagree, and branch supports go below 100, we step over some discrepancies.
A phylogeneticist's standard approach is to blindly believe the high support values, which in this place A. pseudoplatanus as sister to A. velutinum. Which implies that A. pseudoplatanus – assumed to be an autopolyploid – captured not only its plastome from a distant cousin (see next) but also its 35S rDNA cistron from an earlier diverged species (a stem-sect. Acer), because they simply don't fit this alleged sister relationship (Grimm et al. 2007). From an ITS- (ML trees, distance-based Neighbour-nets, in-depth motif analysis of high-divergent length-polymorphic patterns) and morphological point-of-view, A. velutinum forms a lineage with A. heldreichii (not included in Li et al.'s data) and A. trautvetteri: our ‘Group A1’: they share derived and unique to them ITS sequence patterns (genetic ‘synapomorphies’ in a strict Hennigian sense = sufficient criteria for holophyly).
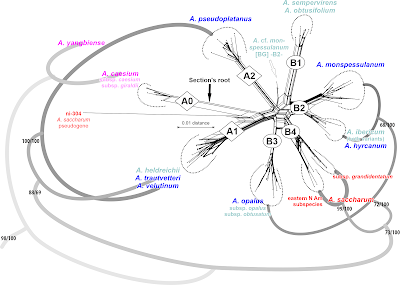 |
| Putatively (quasi-)holophyletic species groups identified within sect. Acer, bracketed by Li et al.'s (2019) ML-inferred subtree. |
While a sister relationship between A. hyrcanum and A. monspessulanum makes sense from a holistic point of view (= ‘Group B2’; see discussion in Grimm et al. 2007), adding A. opalus (‘Group B3’) to that clade may less reflect their respective relation to their North American counterparts (A. saccharum species complex, ‘Group B4’), i.e. a reciprocal holophyly of the western Eurasian (B2+B3) and North American species (B4). It's much more likely due to post-speciation introgression (again, see Grimm et al. 2007, providing hard molecular evidence for introgression between A. monspessulanum, poorly covered in Li et al.'s 2019 data, and A. opalus). Notably, in constrast to what we see in the ITS-based neighbour-net, where I can support and test each topological alternative by shared unique sequence features, including box-like parts of the graph, the branching pattern in Li et al.'s phylogenomic, 500 loci tree, is data-wise trivial: it's just reflecting the overall (total) genetic similarity.
Note the length difference between the ITS-congruent (green) and -incongruent (orange) 500 loci-based edge bundles. Past and ongoing gene flow (in the case of A. opalus and A. monspessulanum-A. hyrcanum) as well as increased shared ancestral similarity (A. pseudoplatanus and A. velutinum have much larger active population sizes than A. trautvetteri, which is generally more distant to any other species of Acer than the other two) can easily overlay and outcompete similarity stemming from an inclusive common origin, an exclusively shared LCA. Especially in generally low-divergent data (note the scale-difference to the ITS-based neighbour-net). Just another little thing, easy to fish for in Li et al.'s data. If one would have looked for it (introgression and hybridisation are not even mentioned in Li et al.'s paper).
Acer pseudoplatanus has not only a different but a very distinct plastome. It carries an H plastome, a lineage that was, according Renner et al. (2008) diverged by the Eocene, in contrast to the C plastomes found any other species of sect. Acer (incl. species not covered by Li et al.) and shared with the (nuclear + plastid) sister clade Pentaphylla-Trifoliata. As a very deeply nested tip within the sect. Acer clade, it would have had to pick this plastome up quite recently. In our 2007 evolutionary scenario, A. pseudoplatanus represent an early if not the earliest diverged species lineage within the sect. Acer core clade (i.e. excl. the Himalayan spp.: A. caesium, A. giraldii, and the new addition, A. yangbiense).
Not impossible, but not probable either: the only other extant species with an H plastome is the distantly related northern North American A. spicatum (sect. Spicata s.str., monotypic; → part 1). More probable is that the undersampling of tips (relying on arboretum specimens of limited representativeness for the species itself) and lack of consistent signal – overlooked substantial inter-gene conflict and/or data noise: all tips have much longer terminal branches than the roots in the according subtrees – triggered local branch attraction and distortion. The blackbox-inferred, untested phylogenomic tree gives us ‘(semi-)false positives’: clades that do not reflect holophyly (monophyly in a strict, Hennigian/cladistic sense).
If the tip relationships in a black-box generated phylogenomic tree differ from those in a broadly sampled, well-studied single- or oligogene analysis, always stick to the latter.
Sure, it's all just little things; details not worth bothering. But can we be sure they are not just the tip of the iceberg? Li and co-workers (and others) pretend to be, with their hardly more than half a page Results. That's the main issue, I have with "big phylogenies" published in Q1-journals, especially when it comes to studying intrageneric evolution. As fully resolved as those phylogenomic trees always are – in case of maple nucleome, even trivial regarding many aspects (uncorrected p distance neighbournet capturing most flat and deep unambiguously supported clades) – as little can the standard blackbox approaches tell us about how a genus evolved and speciated. Obviously the modern-day situation is not the product of a one-splitting-into-two process, strict evolutionary dichotomy. Which we model when using exclusively those fully resolved ML or MSC trees for biogeographic inferences and divergence estimations. The closer we approach evolution's coal-face, the more we should be encouraged to and have to ponder, what they really can tell us about the elephant in the room: the reticulate evolutionary history of a quite ancient – going back some 60 million years, i.e. older than some entire angiosperm families with hundreds and thousands of contemporary species – arborescent plant genus.
In the 3rd and last part of this long-read, we'll look at the worst-possible example of a phylogenomic study on maples, the one of Areces-Berazain et al. (2021), published in just another Q1-journal. A study, where no-one involved in the publication process found it necessary to even look at any earlier paper published on the genus before (phylogenomic or other). And conclude the maple story. Do, what the maple phylogenomicists were either to blind to see, or to shy to point out: juggling the phylogenomic trees against each other and come up with a first evolutionary network for the genus: the Coral of Acer.
Trivia—Peeking behind the curtain: Botanical experts, Systematic Biology, and the impact curse
That Jianhua Li (orcid) didn't want to point to our work is understandable. His earlier papers on the genus are all for the bin, characterised by complete ignorance about the signal in the data he used for his cladograms, and published in journals where he didn't risk to face excruciating reviews. I guess, like for me, his first steps in maple phylogeny, were rock-hard. When I first submitted my Acer phylogeny after finishing my Ph.D. in 2004 to Molecular Phylogenies & Evolution, to the only thing I got was an invitation to review another paper, actually two, just a week after they rejected ours with the reviewers telling us, we should seek help with the phylogenetic analysis etc. I inquired with the editors how somebody who cannot write a paper fit to be published in their journal, should judge the quality of others? I never heard again of MPE and they never heard from me. Win-Win. This early experience of mine with the Beasts lurking in the dark and deep Forest of Reviews is the reason, I never ever wrote a non-comprehensive, non-constructive review, but always criticised and tipped, trying to help the authors to make the most of their data.
Unlike mine (always digging and scratching into the data that produced my trees, and soon, networks), conclusions in Li's papers are drawn based entirely on plastid ‘barcodes’-based cladograms masking substantial ambiguity and lack of discriminate signal. In contrast to our's from 2006–2008 that still, to a good degree stand and were confirmed by new, phylogenomic data sets, they can be easily taken apart. Like Gao et al. (2020) and Areces-Berazain et al. (2021), he largely remained ignorant about what nuclear-plastid incongruence signifies in maples and why it matters, when we weave a story out of the blackboxes' trees. The according sentences in the 2019 paper (which, in all aspects, is much better than anything he published before) read like they have been inserted last-minute, asked for by a reviewer; there are some odd breaks in the narrative throughout the paper. But somehow he managed, as an employee of a respected U.S. botanical research institution (Arnold Arboretum form 1999–2009), to assume a position of fringe power in U.S. systematic botanical circles, which eventually got him the money to go phylogenomic on maples (while I, already in 2008, was cut-off from all lab supplies and sent into exile). Which may explain why such a most interesting tree genus, of high ecological but also economic importance in North America, has been so poorly studied, phylogeny-wise. Because the sole(?!) maple expert of the U.S. prevents it?! He would not have been the first to do so; botany, neo- and palaeo- and especially in the States (having no nobility on their own) can be very feudal. There's a Lordmaster, and this or that plant group remains his (it's still usually a male, despite a majority of botany Ph.D.'s are female) fief for eternity, and he shall not be criticised or challenged, or research done without his grace and permission—personally, I crashed into four of that sort during my active time, starting with my diploma thesis topic (Grimm 1999).
When we submitted our ITS study in 2006 to Systematic Biology (not my idea but we had recruited the right co-author to go for it, and she saw huge potential), Li was one of the reviewers. He wrote an utterly incompetent, unfair and unwitting review. The funniest thing was that he claimed we hadn't taken into account intra-specific and -genomic variation, which had been the reason, we used 606 ITS clones to start with. And the only reason he knew about this phenomenon was because we were the first to document it in that very manuscript and he read it in the abstract (he apparently didn't bother to read the rest of the text). His few but profoundly negative other comments demonstrated he obviously had no idea about phylogenetic methodology (quite common among U.S. systematic [palaeo-]botanists) or could even understand what we did and didn't in our paper. Looking at the many flaws in the papers covered in this longread, I cannot help to wonder, how many of them have been reviewed by Li as one of the “anonymous experts”. And whether that may be the reason, so many misrepresent our 2006–2008 studies.
Rod Page, as editor and equally appauled by using more than trees in a phylogenetic study, didn't see it the same way and was quick to shoot down our 2006 paper for his journal (an article in it can be a career-changer in the WEIRD-world I live). After revision, notably; he couldn't reject it straightaway, I had a too important, from an impact-perspective, corresponing author). But even without Li's help, Page would have turned down the paper: he considered the content not impact-attracting enough for the flagship of phylogenetic journals (current score: 96 citations on GoogleScholar, the paper's data has been frequently used as test-data set for new bioinformatic applications, but is not part of systematic/ phylogenetic botany canon). What editors usually call: “of too narrow interest” or “better fitting for a more specialised journal”.
So, a paper that not only was the first botanical paper to
- show a 600+-tip ML tree computed with a then hardly known programme, RAxML (something Syst. Biol.'s editor and peers were quick to criticise: “untested”; 16212 citations on GoogleScholar for the version launched in that very year; 23623 for RAxML 8, which would become my loyal compagnon for my heretic work), but
- also tested the utility of individual- and species-consensus sequences during tree inference—it increases backbone support by masking terminal noise (a related paper: Stamatakis et al. 2010), and
- introduced bootstrap support networks (following ideas of Holland & Moulton 2003; see also Schliep et al. 2017) to visualise competing branch support and investigate in-depth signal ambiguity in the used data
And dead was a scientific approach, which could have become a new standard and avoid possibly thousands of ‘false positive’ clades in the decade that followed (and until now: Li et al. 2019, Areces-Berazain et al. 2021).
We couldn't even properly upload our 2006 data and results to TreeBase: back then, they could not process trees with such a complex tip set. But there were the one or other bioinformatician back in the days, who wanted to explore further the idea of using ambiguous branch support the way we did in Acer, but there was no fertile ground for that. Biologists loved their trivial stick-graphs and loathed complexity. Many systematicists still do (see this longread's examples). After all, evolution is a simple dichotomous process, isn't it? One Tierchen or Pflänzchen splits up into two and gone it is. That's all, folks!
This very unpleasant "confidential peer review" experience was nontheless the motivation to push the 2008 paper down Syst. Biol.'s throats (currently 89 citations on GoogleScholar), and battle through the paper by Potts et al. (2014, which includes our Acer 606-data as exemplary data set; first version submitted end of 2011, One date that is missing in many scientific publications; 49 on GoogleScholar, 41 on Dimensions). While our 2015 paper on beech (Grimm et al. 2015; probably my very last paper in the flagship journal of phylogenetics, published in my 2###nd### last year in professional science) turned out to be quite a piece-of-cake, review-wise. We still got a nasty one but the new editor was very eager to publish it, smelling [currently 59; 51 on Dimensions] citations rolling in. Only takes a decade to change the tides. By then, they realised such fringe science may bring the minimum impact needed to keep the journal's impact factor > 10 (only 25% of papers in Nature and Science, JIFs > 30, are cited more than 10-times; with >50, >5 “times more than average” in five years, you are already in Dimension's “extremely well-cited” category.
We'll see, if Li's or the other phylogenomic studies published in so-called "Q1 journals" (IF > 3) will at some point break even with those published by someone who has not even been worthy to review them. Would have refused an invitation anyway, if one would flitter in from the likes that published Li et al., Areces-Berazain et al. or Yu et al. I'm out-of-business, I only review interesting studies for no-impact or non-profit journals (when asked, rarely, but still happens). Why? Because the big ones can more easily get “experts”, and many reviews there are just a general waste-of-time: the editor's, when seeking non-biased reviewers, the reviewers', when trying to do a proper job, the authors', when having to decide and deal with the “recommendations” and “suggestions” and trying to rid the final version from all the errors the cheap, India-based proof-setters overlooked, added, or repeatedly ignored. But the insignificant, low-impact ones get those of the victims of those impact-driven cash-cows and “anonymous experts”, and may be thankful for a little bit of help.
Cited literature
Areces-Berazain F, Wang Y, Hinsinger DD, Strijk JS. 2020. Plastome comparative genomics in maples resolves the infrageneric backbone relationships. PeerJ 8:e9483.
Areces-Berazain F, Hinsinger DD, Strijk JS. 2021. Genome-wide supermatrix analyses of maples (Acer, Sapindaceae) reveal recurring inter-continental migration, mass extinction, and rapid lineage divergence. Genomics 113:681–692.
Cardoni S, Piredda R, Denk T, …, Simeone MC. 2022. 5S-IGS rDNA in wind-pollinated trees (Fagus L.) encapsulates 55 million years of reticulate evolution and hybrid origins of modern species. The Plant Journal 109:909–926.
Chen Y, Yang Q, Zhu G. 2003. Acer yangbiense (Aceraceae), a new species from Yunnan, China. Novon 13:296–299.
Delsuc F, Brinkmann H, Philippe H. 2005. Phylogenomics and the reconstruction of the tree of live. Nature Reviews Genetics 6:361–375. [open access via HAL-archives]
Gao J, Liao P-C, Huang B-H, Yu T, Zhang Y-Y, Li J-Q. 2020. Historical biogeography of Acer L. (Sapindaceae): genetic evidence for Out-of-Asia hypothesis with multiple dispersals to North America and Europe. Scientific Reports 10:21178 [e-pub].
Grimm GW. 1999. Phylogenie der Cycadales.Diploma thesis. Eberhard Karls Universität. http://www.palaeogrimm.org/themen/diplomarbeit/inhalt.htm
Grimm GW. 2003. Tracing the mode and speed of intrageneric evolution - a case study of genus Acer L. and Fagus L.D.Sc. Eberhard-Karls University. http://w210.ub.uni-tuebingen.de/dbt/volltexte/2005/1574/
Grimm GW, Renner SS, Stamatakis A, Hemleben V. 2006. A nuclear ribosomal DNA phylogeny of Acer inferred with maximum likelihood, splits graphs, and motif analyses of 606 sequences. Evolutionary Bioinformatics 2:279–294.
Grimm GW, Denk T, Hemleben V. 2007. Evolutionary history and systematic of Acer section Acer - a case study of low-level phylogenetics. Plant Systematics and Evolution 267:215–253.
Grimm GW, Kapli P, Bomfleur B, McLoughlin S, Renner SS. 2015. Using more than the oldest fossils: Dating Osmundaceae with the fossilized birth-death process. Systematic Biology 64:396–405.
Holland B, Moulton V. 2003. Consensus networks: A method for visualising incompatibilities in collections of trees. In: Benson G, and Page R, eds. Algorithms in Bioinformatics: Third International Workshop, WABI, Budapest, Hungary Proceedings. Berlin, Heidelberg, Stuttgart: Springer Verlag, 165–176.
Jiang L, Bao Q, He W, …, Zhang Z-Y. 2021. Phylogeny and biogeography of Fagus (Fagaceae) based on 28 nuclear single/low-copy loci. Journal of Systematics and Evolution doi:10.1111/jse.12695
Li J, Stukel M, Bussies P, Skinner K, …, Swenson NG. 2019. Maple phylogeny and biogeography inferred from phylogenomic data. Journal of Systematics and Evolution 57:594–606.
Potts AJ, Hedderson TA, Grimm GW. 2014. Constructing phylogenies in the presence of intra-individual site polymorphisms (2ISPs) with a focus on the nuclear ribosomal cistron. Systematic Biology 63:1–16.
Renner SS, Grimm GW, Schneeweiss GM, Stuessy TF, Ricklefs RE. 2008. Rooting and dating maples (Acer) with an uncorrelated-rates molecular clock: Implications for North American/Asian disjunctions. Systematic Biology 57:795–808.
Schliep K, Potts AJ, Morrison DA, Grimm GW. 2017. Intertwining phylogenetic trees and networks. Methods in Ecology and Evolution 8:1212–1220.
Scotese CR. 2013. PETM Globe, (PETM_Pgeog_17.kmz, Google Earth format), www.globalgeology.com, PALEOMAP Project, Evanston, IL. ResearchGate. http://dx.doi.org/10.13140/2.1.4535.3601
Stamatakis A, Göker M, Grimm GW. 2010. Maximum likelihood analysis of 3,490 rbcL sequences: Scalability of comprehensive inference versus group-specific taxon sampling. Evolutionary Bioinformatics 6:73–90.
Walker JF, Walker-Hale N, Vargas OM, Larson DA, Stull GW. 2019. Characterizing gene tree conflict in plastome-inferred phylogenies. PeerJ 7:e7747.
Wang W, Chen S, Zhang X. 2020. Complete plastomes of 17 species of maples (Sapindaceae: Acer): comparative analyses and phylogenomic implications. Plant Systematics and Evolution 306:61 [e-pub].
Wheeler WC. 2014. Phyletic groups on networks. Cladistics 40:447–451.
Wolfe JA, Tanai T. 1987. Systematics, phylogeny, and distribution of Acer in the Cenozoic of western North America. Journal of the Faculty of Science, Hokkaido University, Series IV: Geology and Mineralogy 22:1–246.
Xu T, Chen Y, de Jong PC, Oterdoom HJ, Chang C-S. 2008. Acer Linnaeus, Sp. Pl. 2: 1054. 1753. In: Wu Z, Raven PH, and Hong D, eds. Flora of China, Vol 11: Oxalidaceae through Aceraceae. Beijing, St. Louis: Missouri Botanical Garden Press, 516–553.
Yu T, Gao J, Liao P-C, Li J-Q, Ma W-B. 2022. Insights into comparative analyses and phylogenomic implications of Acer (Sapindaceae) inferred from complete chloroplast genomes. Frontiers in Genetics 12:791628 [e-pub].







No comments:
Post a Comment
Enter your comment ...