The maples and me
The maples, Acer, were my first molecular love. I fall in love with them for a couple of reasons, including opportunity. They came into the focus of a molecular project within the framework of a German Science Foundation (DFG) research cluster (SFB = Sonderforschungsbereich) about climate, in which I made my first steps in wet-lab research.Climate, molecular studies on maples? How, and why? are all valid questions.
How? The (blunt) answer is: The DFG, and its peers, are very easily fooled, if you know how. You drop something fashionable at the time (like climate change, this was the SFB's unifying topic) and add something your particular fields (in this case: geological ones mostly) have no idea about (molecular evolution) and they grant and elongate (despite a curious lack of results) a Teilprojekt to study the effect of climate on molecular evolutionary rates in marine (foraminfers) and terrestrial (temperate trees) organisms. To our and the peers defence: although some of the people involved knew it was pretty much science-fiction, there were high-impact papers in the 90s claiming that there is such a correlation based on patchy data. And we can't really say there isn't, because people just stop bothering about it and never looked deep enough to validate or falsify the first results of the last century.
Why maples? Because the botanical garden of the Eberhard Karls Universität in Tübingen has a beautiful arboretum including a lot of (correctly labelled) Acer species from around the world.
You have to milk the cow. And the sort of cheese you're making from the milk matters little in the end. In the long run, our output financed by faithful DFG support was much about the average (according to the DFG's peers). Once, there was an evaluation of the ZMBP, the "Center for Plant Molecular Biology" where we were part of, too (although we could not tap into the big money bags). It turned out that we can write a publication for just 4000 € of research money, while the ZMBP's average was 40,000 €. And ours got as much citations on average.
In addition to my Ph.D. [open access], we actually published a few papers on the maples mentioned in our applications. Evolution-phylogenetic ones, not a single about climate correlation, though (no one really bothered about that anymore, when we applied for our own off-spin projects).
- Grimm GW, Renner SS, Stamatakis A, Hemleben V. 2006. A nuclear ribosomal DNA phylogeny of Acer inferred with maximum likelihood, splits graphs, and motif analyses of 606 sequences. Evolutionary Bioinformatics 2:279–294.—at the time a truly unique paper (see below).
- Grimm GW, Denk T, Hemleben V. 2007. Evolutionary history and systematic of Acer section Acer – a case study of low-level phylogenetics. Plant Systematics and Evolution 267:215–253.—probably one of the best papers, we ever wrote, but there much more to find.
- Renner SS, Beenken L, Grimm GW, Kocyan A, Ricklefs RE. 2007. The evolution of dioecy, heterodichogamy, and labile sex expression in Acer. Evolution 61:2701–2719.—a very quick but timely and obviously still interesing bit of research, much cited.
- Renner SS, Grimm GW, Schneeweiss GM, Stuessy TF, Ricklefs RE. 2008. Rooting and dating maples (Acer) with an uncorrelated-rates molecular clock: Implications for North American/Asian disjunctions. Systematic Biology 57:795–808.—recently copied using a phylogenomic data set (Areces-Bezain et al. 2021), any differing result is because Areces-Bezain et al. naively combined two datasets that are only congruent at (exactly) the section level. More data is not necessarily better (Are complete plastome tree always better...)
- Grimm GW, Denk T. 2014. The Colchic region as refuge for relict tree lineages: cryptic speciation in field maples. Turkish Journal of Botany 38:1050–1066. [PDF]—Introducing A. orthocampestre G.W.Grimm & Denk [EOL/EuroMed/POWO/Wikispecies], the only modern species carrying my author epithet.
The 2006 data matrix with its "six-o-six" sequences has become a standard dataset for bioinformatics playing around with their programmes (hence, the quite different citation numbers on Google Scholar vs. Web of Science) and is probably the first plant phylogenetic study that used an, at the time, obscure programme, RAxML [VI-HPC 2006: 15k+ citations][8 2014: 20k+ citations].
It also is one of the still too few combining classic tree inference with distance-based networks illustrating basic differentiation patterns,...
 |
| Neighbour-net including (A) and excluding (B) sister taxon, tip set reduced to the lesser evolved (shortest root-tip distances) groups elucidating the signal of the deep splits. |
... support consensus networks, ...
... and considering the evolution of prominent sequence motives.
 |
| Sequence evolution of a lineage-diagnostic purine length-polymorphic motif from the 5' end of ITS1 (Grimm et al., 2006, fig. 4). Circles indicate tips (OTUs) that retained the basic motif. |
In short not novel or impressive enough in the end to be published in Systematic Biology in 2006.
[Side note: We had the usual "resubmission encouraged", when the editor considers it a "major revision" (we had one nice and one very nasty review) to keep the time between "submitted" and "accepted" short; but then our RAxML-pimped up re-submission was deemed to be not fancy enough. I sometime wonder how my career would have gone, if they'd published the paper, and my first paper would have got hundreds of citations when RAxML took off soon after, since one always cites another study using the same novel programme and picks those in the high-impact journals). Editors and peers have too much power in science. I got my maple Syst. Biol. co-authorship two years later (Renner et al. 2008)—it was a bit of revenge to submit our plastid analysis there.]
Evolutionary Bioinformatics was an open access journal just launched by Libertas Academia, then a society with similar aims than Public Library of Science or Frontiers in... (now bought off by SAGE publishing, quality has gone downhill), and the paper rushed through peer review (both review reports were more insightful than those we got for Syst. Biol.; for a starter, reviewers and editor bothered to read the entire paper before writing their reports)
In addition, my Acer ITS data sets were re-used for these two methodological papers, in which we propose methods to extract information from polymorphic (within the individual or a taxon in general) basepairs — intra-individual (intra-genomic) variation.
- Göker M, Grimm GW. 2008. General functions to transform associate data to host data, and their use in phylogenetic inference from sequences with intra-individual variability. BMC Evolutionary Biology 8:86.
- Potts AJ, Hedderson TA, Grimm GW. 2014. Constructing phylogenies in the presence of intra-individual site polymorphisms (2ISPs) with a focus on the nuclear ribosomal cistron. Systematic Biology 63:1–16.—once you have one paper in Syst. Biol. or similar high-impact journals attracting some citations...branding shouldn't be but is most important in scientific publishing.
We would have done much more, but then came the year 2008, in which I was forced to leave Germany to get "international experience", according to the DFG (who at the same time congratulated me to our splendid project output). So, I lost my technician, the late Karin Stögerer, with her magic hands that would get a PCR product out of everything, and had arcane knowledge to make our faithful stem of competent E. coli to accept nearly every insert we fed them (she always called me "her post-doc", of course). And our well-equipped lab Ob dem Himmelreich; this was really the street's name: Upon the heavenly realm. It was a fitting one.
Evicted from scientific paradise, I started to roam the wastelands like many other young researchers before and after me. I was lucky (thanks to certain well-meaning collaborators) to directly be offered a chair (to seat on, not to lecture) and computer in the Paleobotan of the Naturhistoriska riksmuseet. A very fine place to work (back then, in all aspects, nice surrounding, nice colleagues), but without the means needed to do any further molecular research.
And the first victim of my forced "internationalisation" where my beloved maples. I tried from time to time to get others interested in them, taking up where we stopped, but didn't succeed. The interested had no extra money to spend, the very few with money to waste feared the complexity.
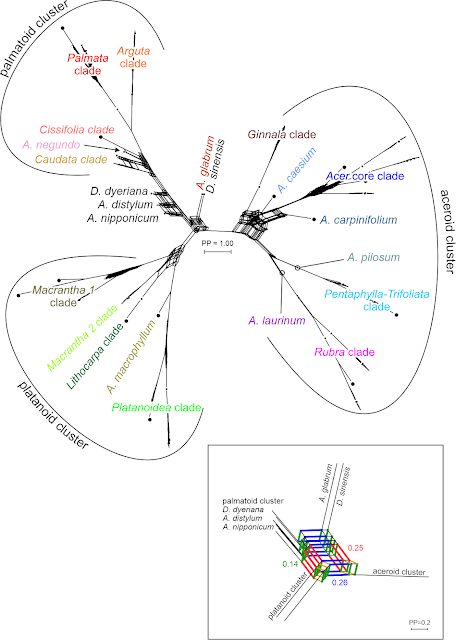
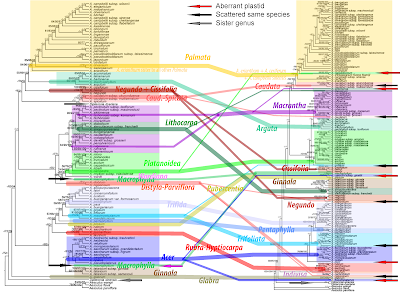
Most recently, they got renewed attention and there are now dozens of complete plastomes, nearly all from China, hence, currently useless to draw any further conclusions about the evolution and biogeographic history of maples. And one new nuclear-based species tree, and a phylogenomic tree riddled by branching artefacts:
- Li et al. (2019), using phylogenomic nuclear gene set – utterly boring paper, poor documentation, pointless discussion evading all critical questions but nice data and a comprehensive sample [Li's negative and profoundly incompetent review – being the only other although very inexperienced researcher working back than on maples, sitting at Harvard Botanical Garden, hence, direct access to many species, published since one pointless cladogram after the other while still ignoring everyhing that makes Acer interesting – was the reason Page had us going into resubmission: the review process should not be "confidential".]
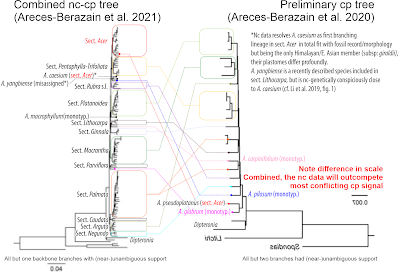
- Areces-Berazain et al. (2021), using a combined chloroplast-nuclear phylogenomic tree. Their sample is more global than in the standard sino-centric complete plastome papers (Wang et al. 2020; Yu et al. 2020) More precisily, it's an ITS-5' ETS tree with a few taxa somewhat displaced (cf. Li et al.'s tree and ours from 2006) because of their highly conflicting complete plastome signatures (long-known: Renner et al. 2008). Interesting is, that a journal like Genomics with an JIF > 5, doesn't even bother to look up what the same authors published the year before in PeerJ: a complete plastome-based tree with much fewer tips, but unambiguously supported clades in general conflict with their 2021 "combined" tree.
What you can do given access to material, resources and/or man-power
Here are some threads that would be worth pursuing and closing up. Not a work for the we-sequence-whatever-we-get-without-thinking-about-it-and-just-dump-it-into-the-blackbox (e.g. Li et al. 2019, Wang et al. 2020, Areces-Bezain et al. 2021; some more examples), but for actual researchers that are interested in how speciation works in extratropical trees and like a bit of a challenge. The former can't deal with it, because what you will need to look into this interesting stuff is material from the wild, more than a single individual per species (all of controlled and informed provenance), and requires a bit more than just inferring data-naive but fully resolved trees.The superspecies and microspecies of Acer sect. Acer
When you look at our 2007 PSE paper, you may (not wrongly) think that the phylogeny of this section is resolved and there is little to find when digging deeper (feel free to check out the according terminal subtrees in the data-intensive but tip-restricted phylogenomic studies mentioned above).But what we found and showed is just the tip of the iceberg. Below a doodle (I made over a decade ago) showing the ITS gene pools in the two complex species of sect. Acer:
- Acer monspessulanum s.l., which includes not only the typical "subspecies" of the western Mediterranean but since the 60s/70s, when lumping became fashionable, a couple of (micro)species, now all treated as subspecies, from the Asia Minor and the Orient, few of which have ever been sequenced or genotyped.
- Acer hyrcanum s.l., which was a taxonomic battleground a bit longer and a close relative of A. monspessulanum (genetically, and morphologically); possibly less complex, but also severely understudied.
- Acer ibericum has two different ITS variants, which may be directly derived from those in A. hyrcanum and local microspecies of A. monspessulanum: evidence for a hybrid origin?
- Acer tauricolum has the genetics of a F1 hybrid, but how's the situation in A. stevenii, the Crimean microspecies (the Crimean peninsula has quite a few intermediate trees, also in other genera, e.g. Fagus taurica, a possible hybrid between F. sylvatica s.str. and F. orientalis s.strictissimo).
- The Bulgarian individual of A. monspessulanum may well be the representative of another ancient (pseudo?-)cryptic species, the last survivor of the source from which the actual A. monspessulanum evolved, like the A. orthocampestre we digged out in the Transcaucasus and described in 2014.
- And there are a lot of interesting blanks. More or less stable morphotypes accepted in local floras but never studied genetically and in the larger context but in areas that are renown for their living fossils ("Tertiary relicts") and are biogeographic cross-roads between farther Asia and Europe.
 |
| Genetic links seen in our very preliminary, unpublished 5S-IGS data |
I just love the 5S-IGS. Pity, I never got the research money to analyse it.
[In case you are interested in the unpublished data, just drop me a note, I should have it somewhere. If you have just money, high-throughput sequencing is an alternative to time-/labour-intensive cloning. Another easy pick for those with funds and access to fresh material would be high-density NGS-SNP studies like the A. Hipp and colleagues did for oaks, which are, data-wise, a harder foe (McVay et al. 2017; Hipp et al. 2019, 2020).]
The plastids, which can be expected to follow more strictly geography, have never been studied to a sufficient detail, i.e. using more than a single individual per species/taxon, and using georeferenced material from natural stands. Being interested in species evolution and putting up first frameworks, we never bothered about them (the easy-to-access markers back then, mostly trnL intron and trnL-trnF spacer lacked information and why study maternal inheritance, when you can have both parental signatures in the much more informative ITS?)
But when you want to put the whole thing in a biogeographic historical framework, you'll fancy a different point of view. One probably doesn't need to dig deep, the easy-to-sequence trnH-psbA spacer may do (it worked wonders for Mediterranean oaks: Simeone et al. 2016, 2018; Vitelli et al. 2017). While often claimed, to study evolution at and below the species level, you usually don't need complete plastomes, only the bits of them that provide the crucial signals (compare e.g. the plastid tree in the supplement of Gao et al., 2020, essentially a trnH-psbA tree, with the tree they published a year later using the complete plastomes, Yu et al. 2021).
Studying the A. monspessulanum-hyrcanum species aggregate will be a tricky but rewarding model case for how extratropical trees speciate. You wouldn't want ot stop with just genetics. Correlating the genetic patterns with morphological clines and devides (or in-depth morphometrics) will give you unique insights about molecular-morphological differentation—what makes a species a species. A lot is done with the herby stuff, but trees outside the tropics are poorly studied regarding evolutionary pathways and patterns and speciation processes. But there is a near-infinite amount of other data: maples are prime research objects in forest sciences, chemo-physiology and other biological fields. Last but not least, one could do spatial distribution modelling and assess climate niches (using 'Köppen signatures', Denk et al. 2013, Bouchal et al. 2018; or 'Köppen profiles'; Grímsson et al. 2018) to see how the genetic-morphological differentiation (nuclear vs. plastid) fits in. And fits with the rugged topography and complex topographic history of the mid-latitudes in western Eurasia. Acer has a fossil record in the region (unfortunately the only expert on these fossils, Harald Walther already died, so one has to start from the material in the collections).
Evolution of Acer sect. Platanoidea
The A. monspessulanum-hyrcanum species aggregate is not the only puzzle to be solved. According to current checklists Acer cappadocicum includes several subspecies, one of which is obviously a species on its own right: the southern Italian Acer lobelii. When we travelled north-eastern Turkey hunting for oaks, we also crossed some very particular and remote populations of subsp./variant divergens. We included samples of both, cappadocicum s.str. and divergens in our ITS studies, and found already some puzzling divergence and polymorphic patterns worth further exploration and geographic mapping.
Their East Asian counterparts, now adressed as subspp. of cappadocicum, are most likely sisters but evolved under different climatic-topographic circumstances. My good friend and former colleague Thomas Denk at the Naturhistoriska riksmuseet in Stockholm, collected more material in southern Italy when hunting for oaks, which we processed in Tübingen in our last days. The data has never been published (again: drop me a note if you need it), but confirmed our early suspicions. Acer cappadocicum s.l. is a super-species or species aggregate and may show very complex differentiation patterns, and not necessarily congruent ones in the nucleome and plastome (the latter is, again, a pretty blank, only the Chinese spp. have been attacked, and only very superficially).
Getting interesting material from the wild can be tricky, but over the years, I was contacted more than once by e.g. researchers in Iran searching for a knowledged molecular lab to collaborate with to study maple (two in the last year). And also in Turkey, Libanon etc. you can find forest botanists interested in maples that may get you the difficult to access material (see e.g. our papers on oaks). Ideally, you find Chinese collaborators. While they are apparently often not allowed to study non-Chinese species on their own (a new funding policy?!), hence, cannot include a single non-Chinese sample even when studying the section (Yu et al. 2020), they have the resources and workforce to do so. Providing them with the proper material, you get most spectacular data sets (see e.g. Yan et al. 2019; Jiang et al. 2021, see supplement to Cardoni et al. 2021 for a proper analysis of their data; Zhou et al. 2021) Reach out and come together! The ones with the money and technology (like Chinese genetics labs), and those with access to the interesting wild stands (e.g. western Eurasian taxonomists, field ecologists or forest biologists).
Why focussing these two sections? Because they are the only truely trans-continental ones that produced a notable amount of distinct species and morpho-genotypes, and are currently under-taxonmised (in contrast to mainly East Asian section Macrantha and purely East Asian sections Pentaphylla-Trifida and Palmata). China may or may not be the cradle of angiosperms, but it even if it hosts most of the accepted species, it only marginally touches the evolutionary interesting ones (like A. mono complex and A. caesium). The treasures are hiding in western Eurasia (especially Transcaucasus), the foothills of the Himalayas, maybe Japan (strangely understudied) and the New World (never really studied, people are too focussed on sheer numbers of species, and the Flora of North America lumped many trees). The genus originated in the Arctic, after all.
The deep incongruence, what else is there in the wild?
The paper of Zhou et al. (2021) is a template for maples as well. (Diploid) Herbs with their perennial or short life cycles and tropical, selectively pollinated trees often have largely congruent nuclear and plastid geneaologies save for the one or other 'rogue taxon' jumping from one position to another. Phylogenetically, in the age of Big Data, they are trivial to study (and phylogenetically-scientifically utter bores). In the extratropical trees, I worked with, they never were. In the Fagaceae covered by Zhou et al., we look back on 80 million years of evolution, many of which they swapped plastomes. Maples are no exception from this rule. Here's a doodle I used in talks a decade ago, summarising the results of our 2006–2008 papers.
And here's how it looks like when one would have not naively combined the data, but just pitted the nuclear (ITS) and plastid (mostly trnH-psbA) cladograms (phylograms are really not asked for in plant phylogentic papers, do we fear branch-lengths?) in the supplement to Gao et al. (2020) against each other.
Or if we compare Areces-Berazain et al.'s 2011 "combined" tree with their complete plastome tree published a year earlier.
Now, most plastid data on maples you find in gene banks (this includes a good deal of the data we used for the 2006 and 2008 Renner et al. papers, lack of resources) are from single arboretum species. They are pretty useless for in-depth analysis of nuclear vs. plastid genetic surfaces, which require georeferenced data points (see e.g. Yan et al. 2019). The same holds for the partial and complete plastome data generated in the last years by the American (Li et al.), Chinese (Gao/Yu et al., Wang et al.) and European groups (Areces-Berazain et al.)
There's a lesson to learn from the oaks, the most diverse extratropical tree genus of the Northern Hemipshere.
It would be easy to take the current basis, add crucial missing complete plastomes – for starters: anything from outside China, from wild populations or arboretum individuals with provenance record – and look further into the deep incongruences with up-to-date tools: the still quite fuzzy inter-sectional relationships. Don't be fooled by "it's just one genus": genetically, one section of Acer can have much more to offer than entire families of other eudicot trees (holds for entire Sapindaceae, their genera a nigh-different, impossible to align ITS-wise, which is no biggy for many other tree families). If one would use genetic pairwise distances to define at which point a clade in a tree deserves a certain rank, to make things comparable with other widespread, northern hemispheric, extra-tropical trees, Acer would at least need to be divided into several genera (especially when considering their plastome data as well), like Heenan and Smisson (2013) suggested for Nothofagus s.l. (and despite being well argued, largely ignored by local botanists, as I was told). And some lineages of Acer probably root deeper than genera in other families. Or entire families.
Being extra-tropical, Acer has a splendid fossil record, but there has been no effort among the living to revise it in the light of the nuclear and plastid genealogies. And the only, probably quite biased matrix of Wolfe & Tanai (1987) has never been published, only the parsimony tree of it and the character list (why we always should publish them; and parsimony was back then the only but is a poor choice to study morphological matrices). Due to the amount of fossils this is not an easy task, but probably would consume a Ph.D. student for his/her entire time. So, it's not feasible: also Ph.D. students need papers (golden three) these days also in a cryptic science such as palaeobotany.
But when you get together a group of the right people, it'd be high-impact research:
- molecular, grant-rich for the fancy data,
- field botanists for the morphological coal-face work (I would see to get a botanist, ideally a taxonomist, not a geologist for looking at the fossils), and
- bioinformaticians/R-geeks to wrap it all up and harvest additional information.
High-impact because you not only have deep nuclear-plastid incongruences to show off (maybe a really ancient reticulation here and there, for sure some chloroplast captures, and with the right data you probably can go for coalescent networks instead of trees) but one of the richest fossil records waiting to be plundered for fancy total evidence and fossilized-birth death dating analyses. See e.g. our papers on Osmundaceae (Grimm et al. 2015) and beeches (Renner et al. 2016), and the many deficits of Areces-Berazain et al. (2021), who didn't bother involving a palaeobotanist—why should they, no-one familiar with Earth's history will be asked to review a paper for a journal like Genomics). And not only a fossil here and there, but several lineages in North America, western Eurasia and East Asia. Two very old (and incomprehensive maps) I generated two decades ago (top: Paleocene; bottom: Miocene).
A lot of pinning points (the one that puts together the fossil record here, should definitely be the first author of the main dating paper, it's by far the biggest pile of work).
Even when you just would do a traditional node dating with the oldest representatives of their lineages (after revisiting the fossil record), you just map the result on e.g. Scotese's free-to-download palaeoglobes (The Easter Egg – everyone can [visually] dive into Earth's past) and not-bad (career-wise) journals like Journal of Biogeogragphy or even GBE or New Phytologist may pick up (the latter even published this nonsensical paper: Larson-Johnson 2016, poor data handling and quality, but good story). Genomics, which took Areces-Berazain et al. (2021) fundamentally flawed paper – all analyses rely on a combined tree with branching artefacts inflicted by joining two deep- (cf. Areces-Berazain et al. 2020) and flat-incongruent data sets of strongly differing signal amplitude – has also a impact factor of >5. Imagine, where you could land with a properly designed maple study!
Maples are much more than just one of the many Sapindaceae genera. It's a complex of morphologically coherent (astonishingly well-sorted) samara-bearing lineages of trees that have been around for 70+ million years (at least since the latest Cretaceous). Very unique, easy to spot but with a very puzzling diversity from the highest to the lowest levels not found in any other tree genus of the northen hemisphere.
Other cited references- Areces-Berazain F, Wang Y, Hinsinger DD, Strijk JS. 2020. Plastome comparative genomics in maples resolves the infrageneric backbone relationships. PeerJ 8:e9483.—misleading title, just compare the tree to the follow-up paper in 2021.
- Areces-Berazain F, Hinsinger DD, Strijk JS. 2021. Genome-wide supermatrix analyses of maples (Acer, Sapindaceae) reveal recurring inter-continental migration [cf. Renner et al. 2008], mass extinction, and rapid lineage divergence [if nuclear-plastid incongruence are ignored]. Genomics 113:681–692.—I'll cover this and similarily poorly designed phylogenomic maple papers in another post for my bad science category.
- Bouchal JM, Güner TH, Denk T. 2018. Middle Miocene climate of southwestern Anatolia from multiple botanical proxies. Climates of the Past 14:1427–1440.—includes also maple fossils.
- Cardoni S, Piredda R, Denk T, ..., Simeone MC. 2021. 5S-IGS rDNA in wind-pollinated trees (Fagus L.) encapsulates 55 million years of reticulate evolution and hybrid origins of modern species. The Plant Journal doi:10.1101/2021.1102.1126.433057.—a colourful but still a tip of an iceberg.
- Denk T, Grimm GW. 2010. The oaks of western Eurasia: traditional classifications and evidence from two nuclear markers. Taxon 59:351–366.—now a classic, using 600+ ITS and nearly 1000 5S-IGS sequences; see Hipp et al. (2020) for the oak species tree.
- Denk T, Grimm GW, Grímsson F, Zetter R. 2013. Evidence from "Köppen signatures" of fossil plant assemblages for effective heat transport of Gulf Stream to subarctic North Atlantic during Miocene cooling. Biogeosciences 10:7927–7942.—much underused concept.
- Gao J, Liao P-C, Huang B-H, Yu T, Zhang Y-Y, Li J-Q. 2020. Historical biogeography of Acer L. (Sapindaceae): genetic evidence for Out-of-Asia hypothesis with multiple dispersals to North America and Europe. Scientific Reports 10:21178 [e-pub].—surprising result, given that nearly all included tips were from China; see my comment on the paper's page.
- Grimm GW, Denk T. 2010. The reticulate origin of modern plane trees (Platanus, Platanaceae) - a nuclear marker puzzle. Taxon 59:134–147.—could have been a template for other tree genera but published in the wrong journal.
- Grimm GW, Kapli P, Bomfleur B, McLoughlin S, Renner SS. 2015. Using more than the oldest fossils: Dating Osmundaceae with the fossilized birth-death process. Systematic Biology 64:396–405.—one of a kind, still; demonstrates what can be achieved when people from different backgrounds and expertise pull together.
- Grímsson F, Grimm GW, Potts AJ, Zetter R, Renner SS. 2018. A Winteraceae pollen tetrad from the early Paleocene of western Greenland, and the fossil record of Winteraceae in Laurasia and Gondwana. Journal of Biogeography 45:567–581. [read-only free access]—joining (palaeo)palynology with fancy phylogenetics is a self-runner.
- Heenan PB, Smissen RD. 2013. Revised circumscription of Nothofagus and recognition of the segregate genera Fuscospora, Lophozonia, and Trisyngyne (Nothofagaceae). Phytotaxa 146:1–31.
- Hipp AL, Whittemore AT, Garner M, ..., Cannon CH. 2019. Genomic identity of white oak species in an eastern North American syngameon. Annals of the Missouri Botanical Garden 104:455–477.—a worst case scenario, solved thanks to NGS.
- Hipp AL, Manos PS, Hahn M, ..., Valencia Avalos S. 2020. Genomic landscape of the global oak phylogeny. New Phytologist 226:1198–1212.—Many authors, all provenances, already more citations (currently 80, with a Dimensions "Field Citation Ratio" of 24 (i.e. 24-times more citation than the field's average).
- Jiang L, Bao Q, He W, Fan D-M, ..., Zhang Z-Y. 2021. Phylogeny and biogeography of Fagus (Fagaceae) based on 28 nuclear single/low-copy loci. Journal of Systematics and Evolution doi:10.1111/jse.12695.—the most interesting data set generated by other people, I ever looked at; a fine example for why Chinese need out-of-P.R.C. collaborators (even Chinese tree species cannot be understood without non-Chinese material). Also a nice example for data- and tree-naivity in the era of Big Data and phylogenomics. None of the authors (or reviewers, or editors) realised what their data actually showed (cf. Cardoni et al. 2021; A fully resolved, and perfectly misleading, species tree). Non-trivial datasets require to do more than just the standard analyses.
- Larson-Johnson K. 2016. Phylogenetic investigation of the complex evolutionary history of dispersal mode and diversification rates across living and fossil Fagales. New Phytologist 209:418–435.—examplifying what can be achieved using fancy methods and poor data, given the right publication environment (see Acknowledgement section of the paper).
- Li J, Stukel M, Bussies P, Skinner K, ..., Swenson NG. 2019. Maple phylogeny and biogeography inferred from phylogenomic data. Journal of Systematics and Evolution 57:594–606.—utterly traditional and boring; the conclusions are pretentious, but nice data and sample.
- McVay JD, Hipp AL, Manos PS. 2017. A genetic legacy of introgression confounds phylogeny and biogeography in oaks. Proceedings of the Royal Society B 284:20170300.—a paper that shows how it's done; a read for anyone working with non-trivial phylogenomic data.
- Piredda R, Grimm GW, Schulze E-D, Denk T, Simeone MC. 2020. High-throughput sequencing of 5S-IGS in oaks: Exploring intragenomic variation and algorithms to recognize target species in pure and mixed samples. Molecular Ecology Resources 21:495–510.—another iceberg's tip.
- Renner SS, Grimm GW, Kapli P, Denk T. 2016. Species relationships and divergence times in beeches: New insights from the inclusion of 53 young and old fossils in a birth-death clock model. Philosophical Transactions of the Royal Society B doi:10.1098/rstb.2015.0135.—despite a lack of molecular data (failing resources), our results still stand (compare them with Jiang et al.'s 2021).
- Simeone MC, Grimm GW, Papini A, Vessella F, Cardoni S, Tordoni E, Piredda R, Franc A, Denk T. 2016. Plastome data reveal multiple geographic origins of Quercus Group [now: Section] Ilex. PeerJ 4:e1897.—when monophyletic species share different plastids; I'd love to see a representative of each of our three main lineages included in a global oak plastome genealogy (see also Oak systematics and complete plastome trees).
- Simeone MC, Cardoni S, Piredda R, Imperatori F, Avishai M, Grimm GW, Denk T. 2018. Comparative systematics and phylogeography of Quercus Section Cerris in western Eurasia: inferences from plastid and nuclear DNA variation. PeerJ 6:e5793 [e-pub].—ups, we did it again.
- Vitelli M, Vessella F, Cardoni S, Pollegioni P, Denk T, Grimm GW, Simeone MC. 2017. Phylogeographic structuring of plastome diversity in Mediterranean oaks (Quercus Group Ilex, Fagaceae). Tree Genetics and Genomes 13:3 [e-Pub].—Simeone et al. (2016), extended version (see also: Comparing neighbour-nets and PCA graphs – the example of Mediterranean oaks @ Genealogical World of Phylogenetic Networks)
- Wang W, Chen S, Zhang X. 2020. Complete plastomes of 17 species [aka individuals] of maples (Sapindaceae: Acer): comparative analyses and phylogenomic implications. Plant Systematics and Evolution 306:61 [e-pub].—one of now many similar studies that are mostly for the bin; don't go for "species", go for provenances when starting to dip into the plastids of extra-tropical tree genera!
- Wolfe JA, Tanai T. 1987. Systematics, phylogeny, and distribution of Acer in the Cenozoic of western North America. Journal of the Faculty of Science, Hokkaido University, Series IV: Geology and Mineralogy 22:1–246.—it's a shame that their morphological matrix has been lost, even if biased, it would have been an asset for any molecular study.
- Yan M, Liu R, Li Y, Hipp AL, Deng M, Xiong Y. 2019. Ancient events and climate adaptive capacity shaped distinct chloroplast genetic structure in the oak lineages. BMC Evolutionary Biology 19:202 [e-pub].—perfect example for the beauty of georeferenced plastid data points, when studying extra-tropical trees at a global scale.
- Yu T, Gao J, Huang B-H, Dayananda B, Ma W-B, Zhang Y-Y, Liao P-C, Li J-Q. 2020. Comparative plastome analyses and phylogenetic applications of the [Chinese spp. of] Acer Section Platanoidea. Forests [an MDPI journal, so probably not reviewed at all] 11:462 [e-pub].—Like Wang et al. (2020), a waste of bytes.
- Yu T, Gao J, Liao P-C, Li J-Q, Ma W-B. 2021. Insights into comparative analyses and phylogenomic implications of Acer (Sapindaceae) inferred from complete chloroplast genomes. Frontiers in Genetics doi:10.3389/fgene.2021.791628.—example for how P.R.C. (being now probably the world biggest scientific paper producer) bias has replaced U.S. bias (the long-standing #1) compromising peer review in scientific publishing (see also Peer review transparency reveals scientific provincialism).
- Zhou B-F, Yuan S, Crowl A, Liang Y-Y, Shi Y, Chen X-Y, An Q-Q, Kang M, Manos P, Wang B. 2021. Evolutionary dynamics driving continental radiations of Fagaceae forests across the Northern Hemisphere. ResearchSquare doi:10.21203/rs.21203.rs-968321/v968321.—currently only a preprint, but Nature Communications' editors would be fools to not take it: a truly supermatrix study, and not combining but separating the nuclear (2000+ gene regions) and complete plastome data (100+ tip set covering all major Fagaceae lineages). Also a demonstration for what Chinese researchers gain by teaming up with non-Chinese having access to the right material.






No comments:
Post a Comment
Enter your comment ...