The nice thing about huge datasets is that they can give quick results, often trivial to interpret. In phylogenomics: a fully resolved, unambigously supported phylogenetic tree. The not-so-nice thing is that downstream analyses using these fully resolved trees, such as ancestral area analyses, may be utter nonsense because the experimental set-up was fundamentally flawed to start with. A post-review of Areces-Berazain et al. (2021), including the results from Li et al. (2019) and Yu et al. (2022).
Forword: This is even for my posts a pretty long text but there so much crap (sorry, but there's no better word for this) published in the context of biogeographic studies, it requires both general and more specific critiques. The recent phylogenomic studies on Acer (Li et al. 2019, Areces-Berazain et al. 2021, Yu et al. 2022), all published in legit Q1-journals, exemplarily demonstrate the utter failure of the peer-review when it comes to such research. Maples are one of the few cases, where we can rely on early gene-poor but well-sampled phylogenies and have a huge fossil record at hand (needs to be revisisted but still) covering >60 million years of evolution across the entire Northern Hemisphere. Major (and common) errors are highlighted in bold and dark red. Interspersed into the critique are many research tips. And, as usual, the end of the post is profoundly positive by giving a general overview how-to-analyse the new wealth of phylogenomic data in the context of biogeographic histories to get studies that fall in the category of research rather than waste. Also as usual, all pics can be clicked to enlarge.
When I first got pointed to the Areces-Berazain et al. (2021) “supermatrix analysis”, I didn't bother to give it a proper look. To somebody who worked a decade ago on maple phylogenies, and has gone network-thinking, already the abstract was somniferously boring.
It starts with the usual commonplaces, we have to put into a paper to make it sound important.
Acer (Sapindaceae) is an exceptional study system for understanding the evolutionary history, divergence, and assembly of broad-leaved deciduous forests at higher latitudes. Maples stand out due to their high diversity, disjunct distribution pattern across the northern continents, and rich fossil record dating back to the Paleocene.
Maybe even latest Cretaceous, but that's just a few more million years. [PS timetree.org gives you a nice overview about hilariously too young estimates, obtained in ignorance of the fossil record: choose Acer as "Taxon 1" and Dipteronia and "Taxon 2".]
Using a genome-wide supermatrix combining [→ Error #1] plastomes and nuclear sequences (~585 kb) for 110 Acer taxa, we built a robust time-calibrated hypothesis investigating the evolution of maples, inferring ancestral ranges, reconstructing diversification rates over time, and exploring the impact of mass-extinction on lineage accumulation.
Very boring. 585.000 basepairs + 110 tips = trivial, fully resolved tree. Could have been interesting, if the data would have not been combined but compared.
Contrary to fossil evidence, our results indicate Acer first originated in the (north)eastern Palearctic region [→ Error #2], which acted as a source for recurring outward migration [→ Error #3].
East Asian origin is trivial given the used topology: a quick tweet with pic. A north-eastern Asian, more precisely Beringian origin (see Denk & Grimm 2009, for beech; a genus not rarely associated with maples in fossil lagerstätten) – not inferrable using Areces-Berazain et al.'s biogeographic scoring – would also not have been really “contrary to the fossil record”: in the Paleocene, maples were distributed from eastern Siberia/Kamchatka (i.e. north-eastern Asia) via Alaska to Greenland and Spitsbergen.
Warm conditions favored rapid Eocene-onward divergence, but ranges and diversity declined extensively as a result of the Plio-Pleistocene glacial cycles.
Trivial for plants in general, but probably wrong for the most widespread and dominant Acer spp. and lineages. Maples are profoundly temperate, the don't like it too cold but also not too warm, hence, are absent from e.g. Central Europe during the Paleocene-Eocene greenhouse phase, when global temperatures >10 °C higher than today (an old classic: Zachos et al. 2001) but can be found on, e.g. Spitsbergen at the time.
These signals in genome-wide sequence data corroborate paleobotanical evidence for other major woody northtemperate groups, highlighting the significant (disparate) impact of climatic changes on the evolution, composition, and distribution of the vegetation in the northern hemisphere.
Great (yawn). Concrete details would have been more interesting: Other “major woody northtemperate groups” are for instance the Fagales, specifically the Fagaceae. And there's one thing they are notoriously known for: the decoupling of plastid signatures from speciation processes, long-known but most recently confirmed using complete plastomes and a set of 2000+ nuclear genes (Zhou et al. 2021). The nucleome informs the species tree, the plastome reflects the biogeographic history. In Acer, this is probably the case, too. Long-known (Grimm et al. 2006, and literature cited therein; Renner et al. 2007, 2008): while major infrageneric lineages, currently accepted sections, show a generally high nuclear (all) and plastid (most) coherence, the intersectional (and intrasectional, as far as studied) relationships are in stark contrast. Especially when using phylogenomic data—Li et al. (2019), 200+ nuclear genes, vs. Areces-Berazain et al. (2020), Wang et al. (2020; Are complete plastome trees...), Yu et al. (2022) using complete chloroplast genomes (further details will be in next post on maples: Big Data = No Brain?)
Which brings us to:
Error #1: Combining two conflicting trees into an artificial one
Why would you “combin[e] plastomes and nuclear sequences”, when it's known that their genealogies are incongruent regarding deep relationships? ~150kp, the usual size of a complete plastome, should be more than enough for a nicely resolved dated tree needed for an explicit biogeographic inference. It's also common knowledge that plastid signatures show a better geographic sorting: a plastid haplotype may not be able to tell me which species I look at, but tells me where a plant/ its mother(s) is (are) from.
In the case of Acer, plastid data gave us a phylogeny, where the mere distribution of taxa makes any explicit biogeographic analysis pointless (Renner et al. 2008).
Why we didn't include one in our 2008 paper: the reconstructed ancestral areas would have been
- trivial for the predominately East Asian clades—the two most speciose comprising a nested North American species;
- ambiguous for the mixed, cross-continental clades and early diverging clades.
Hence, focussed on the (relative) timing of inter-continental splits (we were aware our estimates should not be viewed as highly precise given the data situation).
The obvious nuclear-plastid incongruence (arrows) was the reason why we didn't combine our signal-richer ITS with our signal-poorer plastid data in 2008, when we built a time-calibrated hypothesis to test the maple root and study North American/Asian disjunctions. Any tree inferences (combined-classic or coalescent) has one assumption: the entire data can be explained by the same tree. If you combine two datasets, where one part is the product of a different tree than the other, you'll get an undetermined amount of branching artefacts, or mix of aspect-wise data-constrained branching patterns.
If the amount of phylogenetically sorted alignment patterns is out of balance, the combined tree will tilt to one side. Data gaps can have severe distorting effects. In oligogene datasets, internal signal conflict often expresses itself as high Bayesian posterior probabilities (PP) but low (maximum likelihood) bootstrap (BS) branch support. In multigene, phylogenomic datasets, the one alternative will just outcompete the other. An early, still valid (at all hierachical levels), cautionary tale:
Delsuc F, Brinkmann H, Philippe H. 2005. Phylogenomics and the reconstruction of the tree of live. Nature Reviews Genetics 6:361–375. Open access via HAL.
In Areces-Berazain et al.'s (2021) supermatrix the ~150kbp of plastome have to compete with <10 kbp of multi-copy nuclear-encoded 35S (45S) rDNA, leaving ~425kpb of nuclear gene data added from Li et al. (2019). The plastid data is already outweighted in number of basepairs. Diversity-wise, there are very few plastid regions that can compete with the lineage-diagnostic to high-specific internal (and external) transcribed spacers of the 35S rDNA coding unit (cistron). In the supermatrix, the most tip-discriminative data, the 35S rDNA, is backed up by 425k of fairly congruent genes.
Thus, any combined tree will be inevitably a nuclear-constrained one masking a lot of the conflicting, and biogeographically crucial, plastid differentiation signals.
Not only compared to our 2008 analysis but also when using the authors own complete plastome data (essentially a tip-pruned version of our 2008 plastid tree, details again in upcoming Big Data = No Brain?):
A reading tip for those who have little idea about nuclear-encoded ribosomal DNA, their organisation and evolutionary constraints: an open access review paper fresh from the press authored by my Doktormutter and her polyploidy pals.
Hemleben V, Grierson D, Borisjuk N, Volkov RA, Kovařík A. 2022. Personal perspectives on plant ribosomal RNA genes research: from precursor-rRNA to molecular evolution. Frontiers in Plant Sciences 12:3027 [e-pub].
Error #2: Naïve inferences, using the wrong data
Why is the use of a plastid tree critical in the case of Acer? In contrast to the plastid tree, the East Asian (“eastern Palaearctic” and “Indo-Malay” in Areces-Berazain et al.'s scoring; “eastern Asia” in Li et al. 2019) species are scattered all over the nuclear/ nuclear-constrained tree, and they dominate the (geographically) mixed subtrees:
With so much yellow splattered across the tree, the deeper nodes can only be yellow, too. The same picture can be found in Li et al. (2019, fig. 2; red instead of yellow used for East Asia).
But inferences like can tell us nothing about the origin of a genus. It's a modern-day sample-taxonomic bias.
Primary bias: Overrepresenation of East Asia in modern-day tip sets
One reason for inevitable Out-of-Asia scenarios is that there are two kinds of taxonomic traditions: "splitters" and "lumpers". In Acer, we have the classic situation: the species numbers in the Flora of China (FoC) and the purely/ predominately East Asian lineages – specifically sections Macrantha, Palmata, Trifida (= sect. Oblonga in FoC) – are likely inflated (personal guess: many species described during or following the Cultural Revolution are genetically indistinguishable from a widespread related species). The lists in the Flora of North America can be severly underestimating—some subsp. of A. saccharum are clearly species (e.g. Grimm et al. 2007, and we only scratched the surface). I wouldn't be surprised to find pseudocrypic or even cryptic species in A. glabrum and A. negundo, plastid-wise early isolated lineages (Burning research question: Do all A. glabrum or A. negundo have identical plastomes?) Same for western Eurasia (Searching for a research object? Why not maples!) ITS-wise, we find more divergence in a single individual of the Caucasian A. ibericum – officially only one of many subsp. of A. monspessulanum – than between entire species groups of sections Macrantha, Palmata or Trifida across East Asia (sects Palmata, Trifida) or between East Asia and North America (sect. Macrantha). And both its major ITS-variants are specific, not found in any other subspecies or species of sect. Acer (Grimm et al. 2007). This imbalance in taxonomy is a problem for all phylogenomic studies: they typically only include a single (cultivated, arboretum) individual per accepted species. By focussing on species rather than provenances, we may underrepresent critical geno- (nuclear) or haplotypes (plastid). Which applies also to East Asia (research tip): from a genetic point of view including two individuals from the same widespread species, will give you more interesting and representative data than sequencing five closely related, nearby microspecies. And look out for geographically disjunct subspecies! Such as A. caesium-giraldii.
The second, more biasing reason, East Asian tree spp. are over-respresented in tip sets of northern hemispheric extra-tropical tree genera, is the modern-day climate of the "Golden Plant Triangle": S.W. China (Sichuan, Yunnan), N.W. Vietnam, N. Laos, N.E. Indoburma and the foothill valleys (going down to 1000 m a.s.l.) of the eastern Himalayas—usually undersampled in phylogenomic studies (hard to access, not cultivated in arboreta): Cw – warm temperate (subtropical to mild-temperate) with moist summers and relative dry winters.
 |
| The Golden Plant Triangle and it surroundings. PS Most extra-tropical tree fossils can be found in the upper right corner, not rarely including morphs today only found in the Golden Plant Triangle. Google-kmz high-resolution Köppen map by Rubel et al. (2016), available for download at http://koeppen-geiger.vu-wien.ac.at |
Any non-desert plant/tree can thrive in paradise. And if there's a place, a Eurasian relict lineage can survive the waves of more evolved siblings, cousins or distant relatives invading its niche, it's here. The only other place on the continent is the Caucasus, trickier to reach, and much smaller in area. Not only do the (long) summers have plenty of rain (East Asian Summer Monsoon, up to > 10,000 mm/a), the winters are rather dry, which is important at higher altitudes for trees: sudden cold + humidity = frost damage (e.g. collapsed vessels). Moreover, the steep topography provides niches for all temperature preferences: Subtropical Cwa in the lowlands, changing into fully temperate Cwb at mid-altitudes and snow-rich, boreal (cold-temperate) Dwb further up (see Subtropical ⊊ warm temperate for an overview of terms and climate classification systems). In Central China, it's equally lush, only perhumid: Cfa → Cfb → Dfb. [Reading tip: Unfamiliar with Köppen's climate zones? Check out my "Köppen-Geiger" category.]
For instance, for the Shennongjia Forestry district, Zhu & Song (1999) recorded 17(!) maple species, covering sections Caudata, Ginnala, Macrantha, Palmata, Platanoidea, Trifida (=Pentaphylla), and Trifoliata, at altitudes of 200 (near-tropical, Tann ≥ 17 °C) to 2600 m a.s.l. (cold-temperate, Tann < 5 °C). In the phylogenomic trees, the MRCA (most-recent common ancestor) of the Shennongjia species assemblage would equal the MRCA of the entire genus! It's a place where you can find the end-product of at least 60 million years of maple evolution. Cradle or stomping ground? Not really a question: if they would have been there all the time, we wouldn't find 17 different species of six genetically and morphologically distinct infrageneric lineages carrying at least five main lineages of divergent plastomes.
Phylogenetically, relict lineages, being the leftovers from early radiations, will be scattered around in the so-called "basal", root-proximal part of a tree close to a the corresponding section's root branch. The latter position is also where genetically most distinct members of a lineage have to connect: highly evolved species lineages that underwent severe bottlenecks. When branch-length-aware probabilistic mapping methods are used – currently we fancy Bayesian analysis as implemented in biogeoBEARS, S-DIVA and RASP – the provenances of relicts – genetically more primitive, less evolved living fossils – will have an over-proportional effect on the inferred ancestral area because of their absolute closeness to the inferred MRCAs (most-recent common ancestors).
 |
| The relict problem. |
Evolutionarily, historically, the modern distributions of relict lineages are unrepresentative. Many relict genera, or species groups, in the Golden Plant Triangle had much wider distributions in the past. Those with a fossil record can typically be found in the Palaeogene of north-eastern Asia (Russian Far East, Japan, N.E. Siberia, Kamchatka; e.g. southern East Asian beech species; Denk & Grimm 2009, update in Renner et al. 2016) and even on the other side of the Beringian Land Bridge such as McClain & Manchester's Dipteronia, the stem age prior for all Acer chronograms (Areces-Berazain used it as crown age prior). Others have their oldest fossils in the (then tropical-subtropical) lowlands of Europe (e.g. Engelhardioideae; Rhoiptelea, a living Juglandaceae fossil, is known from the late Cretaceous: Heřmanová et al. 2011; Castanopsis, the subtropical-tropical chestnut sister can be found in the Eocene Baltic Amber: Sadowski et al. 2018). We can assume the same for surviving species lineages: a monotypic, few-species section today (sects Caudata, Distyla, Glabra, Indivisa, Macrophylla, Pubescentia) may only be the surviving tip of an species iceberg. Unless we can estimate their actual original area (fossils!), relict lineages should not even be in any biogeographic inference at all. Probably one reason, so many "explicit" analyses are published in pure-genetic journals such as Genomics (like Areces-Berazain et al. 2021 fundamentally flawed study) and Molecular Phylogenetics and Evolution, or journals with not so stringent peer-review (e.g. Frontiers in Plants Science, Journal of Systematics and Evolution, NPG's Scientific Reports).
Out-of-East-Asia: a nuclear-data sampling artefact
Because of this over-representation, also for Acer any biogeographic analysis using exclusively (Li et al. 2019) or including nuclear data (Areces-Berazain et al. 2021; Gao et al. 2021) must infer or prefer a East Asian origin. It's data-wise trivial – we don't need any algorithm to come to this conclusion – and, most probably, wrong.
Which is even more obvious in Li et al.'s (2019) ancestral area analysis: no plastid data included = everything origins in East Asia. Annoyingly (for biogeographic inference algorithms), the nuclear clades combine a shitload of East Asian species with a single and eastern(!) North American one, i.e. from the exact opposite side of the Northern Hemisphere.
Which lead Li et al. to infer (palaeo-)biogeographically implausible back-and-forth long distance dispersals (BFLDDs):
Quite a jumper, the maples: all started in East Asia (region D), home of the sister species (oldest fossils in B, too, used as age prior for the dating), rushing through western Eurasia (C) and western North America (B) leaving behind no survivors to spawn outposts in eastern North America, just to jump back again and finally radiate in the old home. But why are the geographically constrained plastomes so poorly sorted?
- Explanation one: the BFLDD is a reconstruction artefact having used a wrong model, and it's simple one-time dispersal with further expansion (large population size, quick and barrier-free: little genetic drift) east: Beringian Land Bridge → (north-)western North America → eastern North America. Or west – western Eurasia → North Atlantic Land Bridge → eastern North America – but that route is a) longer and b) would have involved some bottlenecks (topography-related), leading to c) increased genetic drift. The latter would have made the North American endpoint genetically distinct from all its East Asian siblings. Any genetic tree would have placed it as sister to them (inevitable inclade-outclade long-branch attraction).
- Explanation two: the North American species are actually ancient but declining sister lineages of the (still radiating) East Asian species. A few of the East Asians underwent significant bottlenecks, making them genetically most distinct within their section, hence, their lineage (and not the less evolved North American sister) is attracted by the always more distant (in Acer) other sections/ infrageneric groups: local LBA moves them in a wrong, "basal" position. This is particular a problem with phylogenomic data, because few author explore the signal in their gene sample (but see e.g. Walker et al. 2019; Zhou et al. 2022; see also the Why the emperor has no clothes on miniseries on Geneal. World Phylogenet. Networks [Pt. 1: the mighty matK][Pt. 2: a thicket of trees][Pt. 3: conflict or not?])
In case of nuclear-plastid incongruence – a sort of biological industry standard in extra-tropical angiosperm trees – you should always go for the plastomes, when it comes to ancestral area analysis.
Plastids are maternally inherited in far the most angiosperms, and seeds (carrying the mother's plastome and a biparental nucleome) – as a trend – don't travel as far as pollen (carrying the father's nucleome). Unless wind-dispersed, of course, but then there's no nuclear-plastid incongruence beyond resolution issues. Thus, in sexually reproducing plants, they are geographically better constrained. And have been so in the past! Which is a quality, we are looking for, when trying a top-bottom biogeographic analysis. In case of hybridisation, backcrossing with only one donor/parent may quickly homogenise the nuclear gene pool. Asymmetric (unilateral) introgression will have a similar effect. But the original plastid of the first mother will be passed on in the local population, and retained even over millions of years. In extreme (reticulation-prone) cases like the Fagaceae or other extratropical Fagales this leads to ''plastid decoupling'' – individuals, populations, species or entire lineages carrying plastomes that don't match their species or systematic-taxonomic group (cf. Zhou et al. 2022, and literature cited therein). While a nuisance for inferring species trees (Oaks systematics and complete plastome trees), these "wrong" plastids are an asset for biogeographic inferences.
Let's use plastome differentiation common sense on Li et al.'s inferred but improbable BFLDDs.
- Section Negundo—The North American species carries a unique plastome, which diverged from the main bulk of Acer carrying the A to D plastomes (to F, looking at the recent but undersampled complete plastome trees; Areces-Berazain et al. 2020; Wang et al. 2020; Yu et al. 2022) latest at the Eocene-Oligocene boundary (>34 Ma, Renner et al. 2008). Their East Asian siblings (formerly sect. Cissifolia) share the plastome of the (nuclear-inferred) sister lineage, sect. Arguta. A clear case for inter-continental vicariance predating the split into the modern sections. The LCA, the last common ancestor, of section Negundo but also the Arguta-Negundo clade (if genuine) must have been widespread (i.e. including western North America or western Eurasia: fossils essential!), section Arguta evolved in East Asia, hence the shared plastome with sect. Cissifolia: same plastome, same ancestral area.
- Acer/ Pentaphylla-Trifoliata/ Rubra—Again, the pattern is that of a vicariance between eastern North America (sect. Rubra, lineage-unique G plastome) and the putative sister lineages sect. Acer + sects Pentaphylla-Trifoliata (A plastome), originally East Asian. But it's also a sampling artefact: We gave the plastome of sect. Rubra a letter (Renner et al. 2008) because the same plastome is not only found in the Japanese relict sister species of the North American Rubras, A. pycnanthum, but also the (morphologically distinct) oreotropical S.E. Asian A. laurinum, originally included in a monotypic section, Hyptiocarpa (now sect. Rubra, too). Using a genus-representative sample, Li et al. would not have inferred any BFLDD at this node at all. But the inevitable East Asian origin (cf. Areces-Berazain et al. 2021). Plastid-wise, also here the LCA must have been widespread (see below: What plastomes tell us about the biogeographic history of Acer)
- Section Macrantha—The North American A. pensylvanicum has the same plastome than its East Asian sisters, which fall into two main nuclear lineages (Macrantha 1, including A. pensylvanicum, and Macrantha 2 in Grimm et al. 2006, split unambiguously supported in Li et al.'s tree). It hence must represent a relatively recent dispersal from East Asia via western (northern?!) North America into eastern North America. Beringia → lowland Northern North America → eastern North America is a likely route because the plastome of A. pensylvanicum is still very similar to that of two quite hardy North(!)-East Asian species, A. tegmentosum (N.E. China, Korea, Russian Far East) and A. crataegifolium (montaneous W. Japan), forming a species group that could have easily crossed Beringia during e.g. the Mid-Miocene climatic optimum. Maybe even in the Pliocene before the onset of the Pleistocene fluctuations. [Keep in mind: it's very hard for node dating to get sensible estimates for terminal splits with only a few deep age priors.] From it's northern area, the direct ancestor of Acer pensylvanicum escaped the glaciers by just moving south.
Pretty simply, isn't it? In biogeography, grey cells and a little background info still much outcompete even the most fancy computer algorithms (see Denk & Grimm 2009, and the follow-up paper, Renner et al. 2016, for what one can accomplish with a lot of background info; more to come).
 |
| The introductory figure of the Material & Methods section in my Ph.D.thesis (Grimm 2003). An image used to train algorithms for pattern recognition (SZ article about MPI research). While any human, no matter which background, intelligence and age (toddler to senile, so even U.S. presidents) could (and can) readily identify all chairs in the picture, even the best computers and algorithms failed miserably. 20 years later, maybe neuronal networks manage. Maybe they can also come up with non-trivial (and less flawed) biogeographic inferences? |
Telling a story: Historical biogeography feat. Mother Earth
Imaging you are a temperate lineage and just evolved in a cosy place during the Paleocene-Eocene Thermal Maximum, such as Alaska, western North America (≥ 70° N palaeolatitude). Not too hot, summers, long enough to grow a tree. The early Eocene – shown palaeoglobes are available as GoogleEarth kmz files for download from R. Scotese's ResearchGate page – is still a greenhouse: It's a mere cat's jump into East Asia via the Beringian Land Bridge, from there you can directly expand into high-latitude northern Asia: plenty of space, there's also little in the way to stop you going further west. South is trickier. Too much competition and heat in the lowlands. But there are plenty of mountains along the Pacific Fire Ring and in the interior. The massive range expansion will defragment your plastid gene pool, especially if you encounter some bootlenecks in the mountains.
Now Mummy Earth decides to cool down. In the montane areas that's no big deal: you just step down into the lowlands. But your (plastid-wise already different) high-latitude siblings, getting cold roots, may have that idea, too. They go south. Some may have readily escaped to the New World, the land of their ancestors, others start exploring the now cool-enough western shores, which are going to waste because the (sub)tropical natives cannot handle the change. Again others will come in your way, and it's the good old Modern Man meets Neanderthal dilemma: kill (plants replace) or breed (hybridise, intrograde). It just a matter of dices, numbers and niches, which plastome you'd end up with. Some lineages will be completely taken over, only leaving their plastomes as a reminder of their existence (like the ''West-Med'' haplotype in Quercus ilex; Simeone et al. 2016). Polyploidisation may break down fertilisation barriers, with the stabilisiing polyploids randomly picking a plastome and sorting out the other. Or not. While the plastomes get sorted or captured, the nucleomes just homogenise: new species, precursors of future mighty holophyla, will emerge having more than a single evolutionary source. High intra- and low inter-species (-lineage) gene flow will quickly make them genetically unique, irrespective of their provenance.
But there's an escape from the wave: a humongous plateau is lifting up in central Asia forming a lush corridor at its southern flank to go west. And it leads to a pretty place, mountain-ranges-a-plenty with the exactly right niches.
It's getting hot again. Up and north, we go again. But meanwhile those westerners have adapted to the changing conditions, and the corridor is still there. Moreover, the ancestors thriving across the old continent, the New World (from a much-later, human-colonial perspective), have learned new tricks: the first bridge, Beringia, used by the all-ancestors 30 million years earlier, is passable again.
As we approach modern-times, comes the great filter(s). Down-and-south we must go, much will be lost. But there will also be a lot of place for the hardy and flexible ones. The ancients (Ginkgo-paradox: one will always survive), those who never evolved and adapted, have only one hope: Reaching the few places, where it's still like in the beginning.
The mother trees are waiting at their places for the Global Conveyor Belt to jump-start again. Triggering the hot interludes, which will allow them to extend their lineages' ranges once more while spreading locally their plastid legacy, ancient or new, local or from distant shores. With them, a bit of history.
And, mind you: Earth provided your genus with a long-active bridge (''North Atlantic Land Bridge''; Grímsson & Denk 2007), a large, heterogenous and easy-to-roam continent (North America, no latitudinal barriers, old mountain ranges: Rocky Mts and Coast Cordilleras, Appalachians), and the world's biggest archipelago (Canadian Arctic + Greenland), on the other side, too.
What plastomes tell us about the biogeographic history of Acer
By fusing their geographically more informative plastome data with the secondarily homogenised nuclear data – in plants: fast ancient radiations are always accompanied and will trigger latter (intra-lineage) hybridisation and introgression – Areces-Berazain et al. wiped out the most principal geographic splits seen in the genus and ended up with the same nonsensical reconstruction as Li et al. (2019) or Gao et al. (2021). The most obvious misfit regarding Areces-Berazain et al.'s trivial but wrong “eastern Palearctic” (i.e. East Asian) origin, is the plastid of Acer pseudoplatanus, the type species of the genus, hence, section Acer.
A plastome making a huge difference
Acer pseudoplatanus (Group A2), an (assumed) widespread autopolyploid, is morphologically but not genetically highly similar to a group of Euxinian to Hyrcanian species of sect. Acer, A. velutinum and allies (Group A1 in Grimm et al. 2007; A1 because they are genetically less derived than A. pseudoplatanus, both are not part of the most-evolved crown group: B groups). There can be no doubt that all modern sect. Acer species share an inclusive common origin, i.e. constitute a holophyletic – monophyletic fide Hennig – group. The corresponding clade is unambigously supported using Li et al.'s nuclear gene set. Aside from North American (widespread A. saccharum species complex) and western Eurasian species (centre of diversity) they include a remote disjunct and genetically least derived Himalayan-East Asian pair: A. caesium–giraldii (Group A0 in Grimm et al. 2007).
The C plastome is shared with the nuclear-defined sister lineage, the exclusively East Asian Pentaphylla-Trifoliata clade. Already in our 2006 ITS analyses, the two form the crown group of the ''Aceroid cluster''. Sharing a plastome lineage, it's safe to assume that both lineages had the same point of origin (only fossils would be able to tell us where, but morphologically, it'd hard to imagine how their last common ancestor, LCA, could have looked like). Since the lineages are well sorted in their respective C plastomes, it's biologically trivial that the primary divergence was connected to a vicariance event following a dispersal: eastern North America + western Eurasia = sect. Acer, East Asia = sects Pentaphylla-Trifoliata. But this also means that the plastomes of A. pseudoplatanus and A. caesium(?)-giraldii were captured from maple lineages that already existed before the Pentaphylla-Trifoliata and Acer lineages started to radiate. And that both picked them up while migrating into new areas.
Like A. pseudoplatanus, A. giraldii carries a more ancient, earlier diverged plastome – shared with another potential living fossil (A. pilosum, monotypic sect. Pubescentia) and a sympatric, recently described oddball (A. yangbiense) – than the rest of the group (not as ancient as the one A. pseudoplatanus picked up). From the section's nuclear-genetic differentiation patterns, a (high-)montane Himalayan/C. Asian origin with subsequent dispersal along the Hyrcanian-Caucasian-Anatolian Alpine mountain chains into (then already temperate) Europe and then via the North Atlantic Land Bridge (cf. Denk et al. 2011) into North America seems plausible. The alien plastome of A. pseudoplatanus (in German: Berg-Ahorn, mountain maple) has been captured from an already present extinct, ancient European lineage, a long-gone relative of the North American A. glabrum (known as Rocky Mountain Maple in e.g. California): a widespread but isolated species (monotypic sect. Glabra) that has no nuclear or morphological similarity to sect. Acer. Rather than an autopolyploid, A. pseudoplatanus could be a secondarily homogenised allopolyploid of hybrid origin (fishing in its nucleome might bring up some clues). But how does the F plastome in A. giraldii fit in? Acer caesium s.l. may be morphologically and genetically a relict, thriving close to the point of origin but represents an early expansion of sect. Acer eastwards along the Himalayan corridor into C. China, where the ancestor of A. giraldii picked up the plastome of an ancient East Asian lineage of the Platanoid Cluster, with a possible single survivor: A. pilosum. Section Acer's most probable ancestral area is the eastern bit of western Eurasia, the Paratethyan region.
Why we can exclude a cross-Pacific and subsequent North American origin of the section? Left alone the dating results and detailed genetic study of the sections (all B-groups are derived): There are no Western H plastomes in the North American species. If the North American precursor of the western Eurasian section Acer species can pick up a geographically and evolutionary more distant plastome (Eastern H), it should have been possible for the North Americans, too (the distribution areas of A. glabrum, H-plastome and western-most section Acer species A. grandidentatum, have a large overlap).
Ignoring conflicting plastid splits equals ignoring the very biogeographic history of the genus one aims to reconstruct.
The real modern-day biogeographic framework for maples
Unfortunately, Areces-Berazain et al. (2021) discarded their own 2020-results (I asked them why, but so far no answer), but we can match their preliminary tree and the 48-tip complete plastome tree of Yu et al. (2022, fig. 4; their “A. saccharum” is A. saccharinum, there are a few other oddities we'll ignore for now) into our 1 1/2 decade old results.
Topology-wise there are only two slight modifications (we had a pretty clever marker sampling, filling gaps in our preliminary data set, Renner et al. 2007; see also Are complete plastome trees always better...):
- the N. American A. negundo – unique, early diverged plastome – moves a node down;
- and the Ginnala [Research tip: Plastid data on any western Eurasian A. tataricum (sect. Ginnala) still missing] and C plastomes (rest of sect. Acer + sects Pentaphylla-Trifoliata) swap positions.
The related internodes are anyhow short, so these topological modification wouldn't change a lot regarding any dating (we all used the same root constraint, stem age of Dipteronia, only with slightly different values or wrongly linked to a node up).
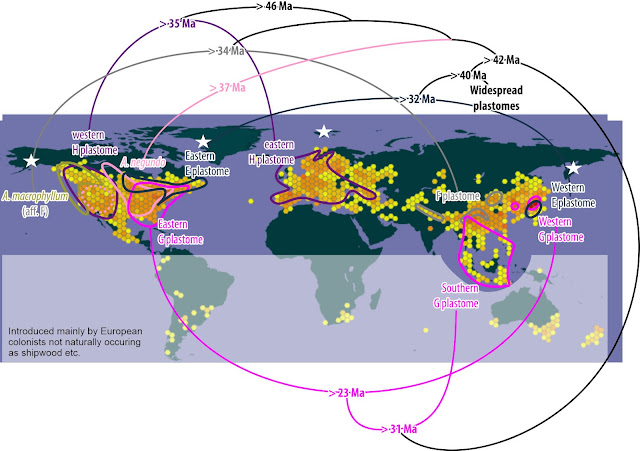 |
| Approximate modern distribution of the first diverging plastid lineages. Background map from GBIF, ranges quickly pulled from GBIF similar open access and open data resources. Divergence ages follow Renner et al. (2008, fig. 3 and table 2). White stars indicate major areas with oldest maple fossils (latest Cretaceous to Paleocene, > 60 Ma) |
Our old results still stand:
- The first geographic split (vicariance) reflected in modern-day data is not intra-East Asia as inferred by Areces-Berazain et al. (2021), Gao et al. (2021) or Li et al. (2019), but between a cross-Atlantic lineage (Clade H: today western N. America [WNA] | western Eurasia [WEA]) and the rest of the genus.
- The next diverging lineage brings us to the Himalayas [HIM] and cold mountain ranges in N. China (part of northern E. Asia [NEA]), Clade F, the plastome lineage also found in East Asian species of sect. Acer (HIM extending to east), ''Aceroid Cluster'' (cf. Grimm et al. 2006) shared with unrelated A. pilosum (NEA), monotypic sect. Pubescentia, ''Platanoid Cluster''. Sister to this lineage is the plastome of A. macrophyllum, a WNA living fossil, nuclear-wise sister of the exclusively (today and in the past!) Eurasian sect. Platanoidea, core clade of the ''Platanoid Cluster''. Leaf-morphologically, A. caesium and A. macrophyllum still resemble the oldest known maple fossils: A. arcticum (p.p., not all fossils stored in collections under this name are maples; T. Denk, pers. comm., early zeroes).
- Soon followed by the North American A. negundo (widespread: WNA + eastern N. America [ENA]), who has a plastome much different from that of the NEA members of sect. Negundo (formerly sect. Cissifolia: carrying the exclusively E. Asian B plastome); and Clade E, another systematically insane plastid lineage with a disjunct cross-Beringian modern distribution (NEA | ENA). Insane, because the three known species with E-type plastomes have nothing in common (morphology or nucleome-wise), except maybe for their general niche: cold-tolerant (hardiness zones 4–7)—A. carpinifolium, oddest possible maple leaves which can be easily confused with those of hornbeams (different family, order, and rosid clade), hence, the name; cold-temperate—A. nipponicum, >1000 m a.s.l. in W. Japan till Hokkaido, where it occurs on 100–800 m a.s.l., Tann ~5–10 °C (Maycock 1994); and cold-loving—A. spicatum, the Canadian ''Mountain Maple'' in southern Canada and northernmost U.S.A., mostly Dfb into Dfc, hardiness zone 2, one of the hardiest living maple species. A relict from the time when the Beringian Land Bridge and northern North America were warm enough for maples?
- Next is again a strongly disjunct S.E. Asian-N.E. Asian-North American lineage including an essential maple species: A. saccharinum, provider of maple syrup (however, not the one figured on the Canadian flag). Unless the ancestor of the modern species was dispersed by Aliens messing with Earth's evolution, the modern distribution can only be a relict distribution. As trivial as the split pattern may look like southern East Asian [SEA]: A. laurinum | { NEA: A. pycnanthum | WNA | ENA: A. saccharinum-rubrum), Rubra (s.str.)-like fossils are not uncommen in western Eurasia. Fossils of A. laurinum (formerly sect. Hyptiocarpa) would probably not be recognised as maples at all, their oreotropical habitats have in addition very poor fossilisation potential.
All this was finished by the onset of the Oligocene, 35 million years ago—like all node datings, the estimates in Renner et al. (2008) should be viewed as absolute mininum ages. See the discussion in the paper for general uncertainty issues.
But these challenging primary geographic patterns, fitting with a High-Arctic ("North American") origin of maples – except for the oreotropical A. laurinum, all these maples thrive in (cold-)temperate conditions – would have led even the most fancy biogeographic inference analysis method to an everything-is-possible area of origin (unless one would explicitly exclude several scenarios prior to analysis, which is the whole point of Ree et al.'s 2005 DEC model). Thus, are missing in Arecese-Berazain et al.'s biased preferred tree used for the ancestral area analysis (or those of others).
Ignorance or negligence? Areces-Berazain et al. cited their own paper with the strongly conflicting and fully resolved plastome tree, ref. 42, only a single time in their discussion: “Our results confirm previous analyses showing the close relationship between sections Trifoliata and Pentaphylla [37,42,76,77] and provide further evidence for merging of the two groups (Fig. 1).” Funny, too, here first-mentioned ref. 76 and 77 are our 2006 paper (Grimm et al. 2006) and the paper of Li et al. (2019; providing the bulk amount of ~425kbp to their tree); both papers showing trees that are substantially incongruent with the referenced plastid-based ones (ref. 37 is Renner et al. 2008).
As in our entirely invented story Historical Biogeography feat. Mother Earth, real plant lineages like maples challenge us with non-trivial, complex information about divergences, range disruptions, and trans-continental migrations. In Acer and other extratropical tree genera possibly pre-dating the formation of modern-day infrageneric lineages (sections). Modern distributions of species forming infrageneric lineages are not representative.
Wouldn't it then be necessary to recruit independent data sets when trying to put up biogeographic scenarios? Especially when the objective is to “... built a robust time-calibrated hypothesis investigating the evolution of maples, inferring ancestral ranges, reconstructing diversification rates over time, and exploring the impact of mass-extinction on lineage accumulation.” Which brings us to:
Error #3: Deus, non-scientia, ex machina
Areces et al.'s Out-of-East-Asia (or Li et al.'s 2019, or Gao et al.'s 2021) is data-/reconstruction-wise as trivial as it is wrong, since they relied on a tree that did not reflect the deepest geographic splits and included many branching artifacts by mixing dominant nuclear signals with locally distorting and conflicting plastid signals (which also means their dating was for the bin: all clock-models assume the data follows the same tree). Plus, by relying exclusively on the inference, they implicitly assume that species' areals don't shift, their modern areals are representative for their lineages, biodiversity centres and biogeographic units are stable over time and that there is no evolutionary reticulation.
But this is not their worst misconception, apparantly shared by their reviewers, editors, and, given how many of similar papers exist, many neontologists.
Feeding a (poorly understood) supermatrix into available black-boxes to infer a dated tree to do an ancestral area analysis is not science but handywork. Doesn't even require special skills, only some time and dedication, the protocols are well-established and most of the work is done by the used softwares. It becomes science only at the moment, we critically investigate the results obtained from the machines. This is where many phylogenomic studies like that of Areces-Berazain et al. fail miserably (There's no need to do what you can't, or more classic approaches: How not to make a phylogeographic study, Trivial but illogical – reconstructing the biogeographic history of the Loranthaceae). And why any study promoting a dated tree and ancestral analysis must be at least reviewed by someone who has an idea about Earth's past. How can one make such a study without having involved any palaeobotanist, palaeoecologist during production or at least review? The authors state it themselves: what makes Acer special, is its “rich fossil record”. Their conclusion:
The results of our analyses do not support the American origin suggested by earlier paleobotanists (Wolfe & Tanai 1987) [brilliant palaeobotanists, but both died before the Molecular Revolution], but instead indicate an East Asian origin for the group [see Error #2]. We argue that the greater abundance of maple fossils in North America is due to a higher sampling intensity in this continent, given by a long history of geological exploration and paleobotanical research [no, it's rather the number of suitable lagerstätten for temperate plants].
But didn't bother to look into “rich fossil record dating back to the Paleocene”. Although:
[from Material and Methods] We performed non-stratified analyses, with and without fossil data, to test for several [more than two?] possible biogeographic scenarios. In the analysis incorporating fossil data, we constrained the ranges at three nodes based on the occurrence of the fossil species we used for calibration. Acer alaskense, A. dettermani [Maple fossils described by Wolfe & Tanai 1987 have nice Wikipedia pages], and Dipteronia brownii McClain & Manchester are from western North America [sic!] and so the respective nodes were ‘fixed’ to include this information. ... [and then in Results] Both [i.e. two], the unconstrained and the fossil-constrained analyses in BioGeoBEARS strongly favored the DEC + J model over the six models tested (Tables S3 and S4). However, the LnL value for this model was considerably lower (ΔAIC =22.4) in the fossil-constrained analysis, thus favoring the unconstrained analysis. This placed the origin of the genus in the Eastern Palearctic region, where it underwent its initial diversification to later [thus, all older N. American fossils must represent extinct, failed sister lineages of Acer] spread to North America, Europe, and south into the Indomalayan region (Fig. 3). [Figure showing the trivial, unconstrained tree; the other “several” possible scenarios not even included in the supplement. Why? Maybe, because their deep nodes would have been a mess.]
Funny. The reconstruction is in conflict with the fossil record but we still use those conflicting fossils to constrain the age of the East Asian MRCAs?
Deus-ex-machina at its best: Thou shall not question thy blackbox. We discard the information provided by the few (three out of hundreds) considered fossils, crucial for providing any age framework, because the (model-calculated) likelihood for a simplifying model used on a fundamentally biased topology inferred from the wrong dataset is considerably lower.
 |
| Not easy to get into North America. Maples were there very early, but without leaving survivors. Annotated Areces-Berazain et al.'s (2021) fossil age priors (see also: The most common errors regarding node dating) |
I argue that when your explicit phylogeographic inference gives you a result that clashes with the unchallenged fossil record, it's because it failed. First-level error(s). The inference has reconstructed ancestral areas that make no sense on the background of the fossil record (cf. cited Boulter et al. 1996) and the tectonic-climatic-vegetation history of the Northern Hemisphere because they are wrong: Unsuitable data were used to infer a chimeric nuclear-plastid tree with branching artefacts and abundance bias towards East Asian tips, which was then ultrametrised and fed to a much-to-simple algorithm/ model relying on a series of implicit assumptions that are not fulfilled.
I further argue that any living palaeobotanist looking into it will confirm that oldest maple fossils (probably stem lineage) found among Acer arcticum are not by accident from the latest Cretaceous/ early(not late!) Paleocene High-Arctic from where the genus – probably after the Paleocene-Eocene Thermal Maximum – dispersed into lower latitudes and nicely temperate mountain niches pretty simultaneously in the New and Old World making ample use of the Beringian (N.E. Asia–western North America; mentioned twice in Areces-Berazain et al. implying it only works in one direction) and North Atlantic Land bridges (eastern North America-Greenland–Europe; mentioned once, referencing a much-cited [copy&pasted], classic paper but ignoring newer literature entirely). By the way, the most important barrier for plant migration but also speciation processes in Eurasia, the Qinghai-Tibetan Plateau, is not mentioned either. Neither is the most important west-east corridor, the southern Himalayan foothills (despite 100+ references).
Morevover, I argue that the modern-day concentration of Acer spp. in East Asia is a Neogene extinction-migration artefact rooting in substantially different patterns, and that the three fossils used as age constraints, not too mention the rest of the fossil record of Acer, better reflect ancestral areas than any modern-day tip set can possibly do. And that using areas like “Palearctis” and “Nearctis” (not too mention “Indo-Malay”, which effectively only hosts a single species of maples: A. laurinum) makes no sense for a genus like Acer.
Finally, I argue that there is no conflict at all.
A perfect fit
Like Li et al. (2019) and Gao et al. (2021), Areces-Berazain et al. (2021) used standard modern-day biogeographic units to investigate the biogeographic history of a genus that was evolved by the Paleocene. At a time when the High-Arctic was warm enough for trees to grow. Including maples.
 |
| Paleocene maples: a clear preference. Background palaeoclimate globe from Robert Scotese's PALEOMAP Project (2000; http://scotese.com/paleocen.htm) |
Which explains the odd pair making up the first diverged plastid lineage, Renner et al.'s ‘Clade H’. The plastid lineage unique to two otherwise unrelated species, a montane North American, phylogenetically isolated species (monotypic sect. Glabra) and a polyploid western Eurasian member of the ubiquitous section Acer directly reflects the genus' Arctic origin – global greenhouse climate, Arctic not covered by ice and fully accessible to trees – with subsequent dispersal into mid-latitude montane areas and temperate niches of North America and western Eurasia and vicariance in the Oligocene, when the Arctic connection between this old North American-western Eurasian lineage broke down.
Now, for those who have no idea about fossil records (even less than me), i.e. neontologists like Li, Areces-Berazain, and Yu et al. or their editor/reviewers, there's always GBIF.
Let's map the modern distribution of the two species carrying the plastomes representing the first geographically isolating lineage of maples (Renner et al.'s Clade H) on the fossil record of Acer as covered in GBIF.
What a coincidence, the earliest diverging plastid lineage is shared by a widespread montane western North American species A. glabrum, and pops up as oddity in an equally widespread, fully temperate western Eurasian species, A. pseudoplatanus. Even the modern ranges of these two species coincide with the abundance of maple fossils. In case of A. glabrum, geographically close to Areces-Berazain et al.'s age priors A. alaskana and A. dettmani. How probable can it be that these plastomes migrated (much later) from East Asia (A. glabrum ~ 50 Ma; A. pseudoplatanus < 20 Ma) and only survived in two unrelated species? We find them, because their carriers stayed pretty much in place. Acer glabrum may be the sole surviving species of the Clade H lineage, the first maples to leave the High-Artic. Acer pseudoplatanus consumed (the rest of) its eastern sister.
But the map shows more: the problem with potential and realised niche. Once introduced into North America by European colonists, A. pseudoplatanus readily naturalised because of the availability of suitable niches. It's pretty naive to assume – as done by unchecked biogeographic inferences – that such invasions don't happen naturally given the right climate-tectonic-geographic framework. Given a couple of million years, the North American A. pseudoplatanus will have different plastomes from their European sisters. Given the already large active population size, the nucleome may take longer to diverge but eventually there will be two distinct species. If the next intelligent species will do maple phylogenetics, their data will be even more puzzling.
Note also the concentration of GBIF-recorded fossils in old montaneous areas, especially when it comes to mid-/low-latitude East Asia (a notable difference to the fossil record of beeches, predominately oceanic). In modern-day low-latitudes, extant Acer spp. are still confined to the mountains. It's trivial that during global greenhouse phases, preserved or not, maples could have survived and radiated mainly in the mountains as well. Remember Clades E, F and G? By the late Eocene (~40 Ma), the first bulk of plastid lineages were diverged. Here's how the maple world could have looked like during the early phase.
Little to say, except that it makes a lot of sense. From it's circum-polar Arctic origin, the Pacific-facing lineage of the genus readily spread along the mountain chains into western North America and North-East Asia (NEA). The lack of connecting mountain chains hindered its Atlantic-facing sister's expansion into, and subsequent radiation in, western Eurasia and Central Asia, although first bridgeheads were established. From which the genus, and the newly forming modern sections, could easily expand in all directions during the cooler Oligocene.
So many fossils to ignore in future phylogeographic analyses
Now, keep in mind that GBIF includes the digitalised or published georeferenced fossils in museums and collections. It's not comprehensive, and may included mislabelled material—like many other records of modern genera, the fossil record of maples is in dire need of revision. But overall, it's a pretty neat map (typical maples are really easy to spot as fossils).
Another (old, not updated) series of maps with fossils for comparison (fossil beech finds included for comparison, updated by our work in progress on beech):
Already this nearly 15 years old compilation (done for a talk, mostly based on the classic papers, I used for my Ph.D.: Walther 1972, Tanai 1983, Wolfe & Tanai 1987, Boulter et al. 1996) has some areas not in the GBIF data (such as Greenland, Alaska, north-eastern Siberia, Kamchatka) but it's probably outdated.
Note that these maps are done by someone with little prior knowledge about (fossil) maples just by seeving through primary palaeobotanical literature (General tip: It helps to be able to read German, otherwise you miss out on very important classic bits). Modern science: getting a supermatrix is no big deal anymore but having somebody tabulating primary literature and mapping the published fossil record...the 100 references of Areces-Berazain et al. (2021) include only a handful of primary palaeobotanical papers, every single one copied from our earlier papers (hence, basis for the my 15-odd years old maps!) But have been quick to dismiss the conflicting fossil record:
Despite a rich fossil record, it is very difficult to place extinct species of Acer in modern sections [not difficult, only a lot of work]. Several authors [cited are Wolfe & Tanai 1978, and Tanai 1983, i.e. one team] have assigned fossil leaves and detached samaras to currently recognized sections, but most of these assignments are not reliable because the fossils lack the diagnostic characteristics of the sections, and can match the morphology of more than one section [cited is a North American master thesis; but see also Boulter et al. 1996 and Walther 1972]. For this reason, we did not include a high number of Acer fossils in our analysis but rather selected two fossil species that can be confidently assigned to extant clades and thus can provide internal calibration points.
Placing fossils is the most basic problem that would need to be solved when one wants to put up better biogeographic analyses. Boulter et al. (1996) drew a pretty compelling picture anyhow, in good fit with our 2008 dating results (cf. Renner et al. 2008). And misplaced or not, they are still maples, and they have been in North America (and western Eurasia!) at a time when the inference only recorded East Asian lineages with one exception: A. glabrum. How plausible is it, that only one of the original North American (old mountain ranges, no west-east barriers = little risk for extinction) maples survived and all others migrated (much later) from East Asia? And not a single one into or from western Eurasia, despite a viable land bridge; thanks to the Gulf Stream, the northern North Atlantic is longer accessible than Beringia during global cooling (cf. Denk et al. 2013)?
The reconstructed origin of each section of Acer(using plastid data!) can only be verified using the fossil record. If there remain conflicts, they have to be studied – is it the molecular data? is the fossil misassigned? – and solved. Waving them away, is not a scientific approach!
By the way, most abundant, and probably best represented, are the maples of Europe not covered in Wolfe's and Tanai's compendia (haven't read the McClain's master thesis). See discussion and references in Grimm et al. (2007) or Grímsson et al. (2020). Grimm et al. (2007) is a classic piece about what palaeo- and neontologists, field morphologists and wet-lab geneticists, can achieve when working together, and with little resources. I'm particularily proud of this paper because it was reviewed in depth by the late Harald Walther [German Wikipedia], the last surviving palaeobotanist who knew his maples very well. He also wrote one of the most pleasant
lines, I ever read in a reviewer's report: “Although I cannot judge the phylogenetic analyses performed in this paper [a combination of tree and network inferences, and sequence motif analysis], I like their results.” Because they fitted so well with his experience from the having studied the fossil record for a long time. As usual, I might add: well-studied fossil records and well-understood genetic data always match up (now looking back on 20+ years as a hinge between neo- and palaeontology; see my GoogleScholar profile and those of my (former) co-authors for dozens of examples).
A common problem with “rich fossil records” (compare Boulter et al. 1996 with recent phylogenomic papers): they are easily in conflict with "explicit" biogeographic inferences, not only because neontologists are shockingly data-/inference-naïve (Errors #1, #2, and #3) but because plant lineages migrate and mingle, and the modern distribution of certain species or entire (genetic) lineages can be unrepresentative, Earth's tectonic and climatic history needs to be considered, and there are too many fossils and too few palaeobotanists. But the when and where, provided by each fossil as direct evidence, is an invalubable puzzle piece.
A few things are obvious, already from this likely incomprehensive and outdated picture:
- The congruence between plastome genealogy and early fossil record of Acer is not a coincidence. Maples are not out-of-East-Asia but High-Arctic in origin. From there, they migrated into the montane areas of lower latitudes simultaneously in the New and Old World and started to radiate: the plastid pools of East Asian, western and eastern North American, and western Eurasian species got gradually separated. First they colonised the mid-latitude mountain range topping the still too hot lowlands, which they could invade in the Oligocene when Earth cooled down to modern-day situation, finally reaching low-latitudes (A. laurinum colonising the Malesian Archipelago). The nucleome-informed species tree masks many of these early radiations, paramount to understand the history and origins of maples in the Northern Hemisphere.
- There must have been several phases of secondary inter-continental migration, starting in the late Eocene/Oligocene (Renner et al. 2008). Some lineages (modern-day sections) went east-west (sect. Caudata: western N. America → E. Asia), others west-east (sect. Macrantha: E. Asia → western N. America † → eastern N. America), both west and east (sect. Acer: Himalayas into W. Eurasia + eastern and western N. America and C. China), or back-and-forth (sect. Platanoidea) after maples went south colonising any available ± temperate niche in the Northern Hemisphere. Cards were re-shuffled, species formed and fused (within sections) during the Miocene high-time, when global temperatures went up again and the Arctic became re-accessible (Mid-Miocene Climate Optimum).
- North American-European migration used the North Atlantic Land Bridge corridor, potentially active for maples since the Paleocene till latest Miocene.
- Likewise, Beringia was a possible corridor during greenhouse phases.
- Cross-Eurasian migration could use two routes. The northern route via southern Siberia along the northern flank of the Paratethys, passable for hardy, (cold)temperate lineages such as sect. Platanoidea p.p. during global greenhouse and intermediate phases; the southern route via the Himalayan corridor (southern foothills), the Iranian, Caucasian, Anatolian mountain chains for the rest (sect. Acer, sect. Palmata) and at any time thanks to its variable topography.
- Intra-continental migration in response to global heating and cooling: North-south in (eastern) North America due to the general lack of west-east strifing barriers. And between northern East Asia (Mongolia, N.E. China, eastern Russia, Korea, Japan), the original and persisting biodiversity hotspot of temperate modern tree genera and home to all main plastid maple lineages, and southern East Asia (rest of China and adjacent India, Burma, Indochina), the modern-day refuge for all relict lineages.
The general patterns are not unlike the beeches (The challenging and puzzling ordinary beech...; with a bit of oak-ness), a temperate climax tree, which, in many fossil floras of western Eurasia, (North-)East Asia and North America, can be found together with maples and also evolved in the late Cretaceous (Grímsson et al. 2016). The earliest record (see also Denk & Grimm 2009; Renner et al. 2016) is puzzlingly analogous: Oldest (stem) fossil is from Wyoming, north-western U.S.A., already back then highlands (even higher than today); the oldest known modern-beech pollen from the Paleocene of N.E. Russia and W. Greenland. The oldest complete modern-day beech is from the Eocene Okanagan highlands of British Columbia (~50 Ma; Manchester & Dillhoff, 2005), another co-incidence (also for beech, explicit biogeographic analysis would promote an out-of-East Asia scenario when using the wrong tree, or method, B. Moore, pers. obs. 2003–2004; that was before Ree et al. opted for a trivial, easy-to-fix example for their DEC model). Already long-dead palaeobotanists noted this pattern, hence, Acer has been a key element of Engler's and Chaney's ''Arcto-Tertiary Hypothesis'', the idea that an originally high-latitude (circumpolar) temperate flora gradually replaced the low-/mid-latitude ''paratropical'' elements during the Cainozoic. See e.g. Grímsson et al. (2015) for a joined palaeobotanist-geneticist discussion of the topic (naturally in a low-impact journal, like most sincere palaeobotanical research). But palaeobotanists, the few that are still alive, are rarely recruited as reviewers for phylogenomic papers, even when those papers claim to have “built a robust time-calibrated hypothesis investigating the evolution of maples, inferring ancestral ranges, reconstructing diversification rates [pretty tricky, when one combines data from to two differently evolving genomes] over time.”
Just with a bit of (fossil) spice, it'd (have) be(en) so easy to make an interesting paper. Reading into the literature about “other major woody northtemperate groups” such as beeches (ecologically more, much fewer spp.) or oaks (ecologically less constrained, even more spp.), could have give some ideas as well.
What Areces-Berazain et al. should have done with their supermatrix
Here's what I would have suggested, if they would have asked me (I give tips for free) or if I would have been a reviewer of their paper. [Which would have been impossible in this case: a) Genomics has a too high impact factor to ask me for a review; b) I would have declined anyway, because I don't work for Elsevier for free. You shouldn't either.]
Basic level analyses
With 585kbp data at hand, you easily get fully resolved trees, you don't need to waste the entire data for a single tree. We only combined nuclear and plastid data in the past because they, on their own, didn't produce enough support: plastid data would stabilise the backbone but fail miserably in the leaves, the nuclear data (back then mostly ITS), would fix that but the deep signals would usually be diffuse. And most reviewers didn't accept any hypothesis if not linked to a clade with BS > 70, PP ~ 1.00. With Big Data and phylogenomics, there's absolutely no point to waste good data by combining it!
- Infer a (simple and/or coalescent) tree based on tip subset of Li et al.'s (2019) nuclear gene data and tangle it against a 35S rDNA-only tree. Use phylograms, to illustrate the divergence level. There may be some incongruences, especially towards the tips, which can be explored. The gene data will be stronger affected by incomplete lineage sorting—are there notable differences between the simple-combined, RAxML/IQTree and coalescent, e.g. ASTRAL, tree? The 35S rDNA data will have superior resolution below the section level because of the variability and information comprised in its transcribed spacers, but the gene tree will be more straightforward regarding inter-sectional, deep splits.
- Infer a complete plastome tree, tangle it against the two nuclear trees. This will allow pinpointing the highly supported topological conflicts, deep (nuclear gene tree) and flat (35S rDNA tree). Note that by using the same tipset for all three trees, you can use the consensus tree approach implemented in SplitsTree (Huson & Bryant 2006) or Phangorn library for R (Schliep et al. 2017) to directly visualise all conflicting branching patterns. A simple tool is also to just make x-y plots of the competing split supports. For differing tip sets (missing data, if you want to include them), use supernetworks. General tip: The most useful partitioning scheme for any complete plastome analysis is the genetic-logical one, especially if even PartitionFinder2 comes up with totally insane ones (genetically and computationally). For our beech complete plastomes, it e.g. suggested >100 partitions. Five or six will do: (i) all 1st + 2nd codon positions, defining the amino acid sequence, thus prone to directed/ positive selection, (ii) 3rd codon position, enriched in synonymous mutations, also may be saturated in some cases, (iii) introns, non-coding but usually more conserved than, (iv) intergenic spacers, (v) structural RNA genes, tRNA and rRNA genes (can be joined or kept apart, will matter little). Check and correct for inversions, don't bother about coding indels. The partioning scheme used by Areces-Berazain et al. (large single copy region—LSC, small single copy region—SSC, inverted repeat region—IR) is baseless; but in those cases where plastomes are biparentally inherited (e.g. certain gymnosperms), comparing the LSC-, SSC- and IR-only trees can be interesting.
- Use your fossil set to date the nuclear gene tree (for fun, you can date the 35S rDNA tree, too, but it's not advisable from a theoretical-methodological point of view), and use the same root constraint to date the plastome tree. This way you get two dated, equally root-tip scaled trees that can be just overlaid. Extract the taxonomic-geographic decoupling events: e.g. for the A. pseudoplatanus case, such a comparison will directly show that the plastome was already diverged before the species evolved. Notable are also situation where geographic (plastid) and species (nuclear) divergences appear to be coeval although the background topology is different. Be wary when using in-tree constraints when dating plastome tree: e.g. the oldest fossil related to A. pseudoplatanus may or may not have carried the A. glabrum-related plastome. Where transferrable, make explicit use of the geographic information provided by fossils. It's illogical to use e.g. North American fossils as age priors for MRCAs reconstructed as East Asian (→ Error #3) and referring to the initial intra-East Asian divergence. If the oldest fossil of a largely East Asian section such as Macrantha is western North American, then the fossil can only represent:
- an extinct sister lineage, hence, informs the section's stem age, or
- the first member of the North American sublineage, i.e. informs the stem age of the according subtree/ tip (i.e. A. pensylvanicum).
- Map your phylogenetic results on the past Earth. E.g. using Scotese's open data palaeogeographic maps (The Easter Egg – everyone can dive into Earth's past). If you feel the urge to do a explicit biogeographic analysis (which I, personally, find superfluous but it's a selling point with reviewers in Q1 or high-impact journals), use biogeographic units that are coherent over time, the time range covered in the dated tree. To infer ancestral areas of lineages, e.g. testing for a North-East Asian vs. western North American origin, the putative candidate ancestral areas need to be scored! Not standard-used ones like ""East Asia"" or ""eastern Palearctic"". The scored regions should be adapted to the current and past distribution patterns. For maples, a natural spatio-temporal concept would be:
- western Eurasia—Europe till Ural/Turgay Strait, Asia Minor, Caucasus into N. Iran (Euxinian-Pontian-Hyrcanian mountain ranges)
- Himalayas and Central Asian mountains [corridor(s)]—Hindukush, Altai, southern Himalayan foothills; a subdivision may helpful for directly discerning northern (cold-temperate, WEA↔CAS↔NEA) vs southern (all others, WEA#harr;HIM#harr;SEA) migration routes
- southern East Asia—S.W./S./C. China (south of the Yellow River), Assam, Burma, Indochina, Taiwan; insular S.E. Asia (Malesia, Philippines) can be included since only hosting one species
- northern East Asia—N./N.E. China (north of the Yellow River), Mongolia, S.E. Siberia, Russian Far East, Korean Peninsula, Japanese Archipelago
- Beringia [past corridor]—N.E. Siberia, Kamchatka, Alaska, Yukon (northwest of Cordilleran main ranges)
- western North America—Cordilleran mountain ranges (Rocky Mts) and western forelands
- eastern North America—everything east of the Rocky Mts, Appalachians
- possibly: northern North America—Canadian interior, N.C. U.S. (today BSk, Dfa, Dfb-climates; easy to access for all temperate maples with a slight global warming)
- northern North Atlantic [past corridor]—N.E. Canada, Greenland, Iceland
 |
| Modern-day. QTP = Qinghai-Tibetan Plateau, major barrier. Red font: extinct ancestral areas. |
| |
 |
| Back-in-time: Eocene, 50 Ma. Major barriers for maple migration in red: Turgay Strait, major sea-way; Paratethys, large-shallow intra-continental marine basin fringed by "paratropical" (extinct climate situation, ≈ hot subtropcial) lowlands bordered by arid climate (Köppen's BS, BW) towards the east (see e.g. Scotese's 2000 maps [early Eocene][late Eocene]). Globes from Denk & Grimm (2009), custom-tilted and provided by R. Blakey (new homepage: Deep Time Maps). |
Using e.g. Scotese's palaeoglobes (or Blakey's) and -climate maps (based on sedimentary etc. indicators), a subsequent feeding the DEC model with no-go-at-time-x-constraints will be easy. Keep in mind: the more well-argued fossil/ abiotic constraints force an explicit biogeographic inference on track, the lesser the risk it derails and strands in out-of-East-Asia or all-is-possible voids. We only do them to push forward a hypothesis, produce a fancy graph for the reviewers and other readers or handle big tip sets, not because they really can test anything. [General warning: Ignore all suggested constraints in papers by Buerki and co-workers, 2011ff.]
When doing biogeographic inferences, avoid mixed tips. Prioritise the actual provenance of the sequenced individual over the cummulative range of the species! It's just a reconstruction, a means to an end, where we use already a lot of implicit assumptions (such as that the modern distribution area is representative, all tips are genetically coherent species, ...) We can afford some explicit ones. Eastern Himalayan species, found at mid- to high altitudes in India, Nepal, Bhutan and Tibet, will typically extend into similar habitats in S.W. China (Sichuan, Yunnan) or even the mountain of C. China (Shaanxi, Hubei, Hunan). They are still primarily Himalayan and should be distinguished from those restricted to subtropical-tropical Assam, northern Indochina, Guangxi, Guangdong, Hainan, Jiangxi, Fujian (= southern East Asia). Vice versa, there are warmth-loving species extending from southern East Asia, where they have their abundance centres, into the relative low-altitude valleys of the Himalayas (India, Bhutan, Tibet) or sea-level, subtropical W. Japan. Which should be contrasted from the typcial, (cold-)temperate northern East Asian species that rarely can be found south of the Yellow River (if, than only towards Gansu and Qinghai) but are common in N.E. China (Liaoning, Jilin, Heilongjiang), Korea, Russian Far East, and extend via mid-/high-altitudes W. Honshu into Hokkaido and Sakhalin. Because of their disparite preferences, a Himalayan, southern East Asian and northern East Asian lineage would have responded differently to past climate fluctuations, which may have left an imprint in their plastome signatures. Like the Gulf Stream heats Europe, the Kuroshio Current ensures snow-rich but relatively mild winters along the north-western North Pacific coast, the more powerful it gets, the easier it becomes to (re-)reach western North America. For some genera, it may be useful to subdivide further North America or western Eurasia. For others, a simpler area scheme may be more appropriate.
Here's a list of useful literature to quickly see the basic distribution patterns of contemporary tree species to make a decision in borderline cases:
- Fang J, Wang Z, Tang Z. 2009. Atlas of Woody Plants in China. Volumes 1 to 3 and index. Beijing: Higher Education Press.
- Thompson RS, Anderson KH, Bartlein PJ. 1999. Atlas of relations between climatic parameters and distributions of important trees and shrubs in North America — Introduction and Conifers. U.S. Geological Survey Professional Paper 1650–A:1–269.
- Thompson RS, Anderson KH, Bartlein PJ. 1999. Atlas of relations between climatic parameters and distribution of important trees and shrubs in North America — Hardwoods. U.S. Geological Survey Professional Paper 1650–B:1–423.
- Thompson RS, Anderson KH, Bartlein PJ. 2001. Atlas of relations between climatic parameters and distributions of important trees and shrubs in North America — Additional conifers, hardwoods, and monocots. U.S. Geological Survey Professional Paper 1650–C:1–386.
- Browicz K, Zieliński J. 1982–1994. Chorology of Trees and Shrubs in South-West Asia and Adjacent Regions. 10 vols. Poznan: Polish Scientific Publishers.
Naturally, checking out GBIF will not hurt either. Needs careful post-processing for invasive and ornamental species. GBIF has no filters for natural vs anthropogenic distribution.
And even without a palaeobotanist on bord, it would have been an interesting read.
Level 2 onwards
Of course, one can do much more with such data.
- Explore gene conflict and support in the nuclear gene matrix. Many of these randomly collected genes can be bare of discriminative signal, while others may be high-informative. The limitation of large gene samples covering few individuals is obvious in maples, when comparing the sect. Acer and sect. Platanoidea subtrees in the trees of Li et al. (2019) and Areces-Berazain et al. (2021) with our earlier in-depth results relying on comprehensive samples covering both intra- and inter-specific variation. If there was reticulation in the past (introgression, hybridisation, polyploidisation), some genes may actually reflect different evolutionary pathways. Easy to spot is secondary contact due to inter-continental migration (see e.g. Cardoni et al.'s 2021 re-analysis of Jiang et al.'s 28-gene data on beech). Note that the now fashionable coalescent tree inference only compensate for incomplete lineage sorting; they cannot identify actual reticulation. The now kind-of-standard cloudograms are also not overly informative in this regard: each single gene tree may have a lot of random splits, not supported by any alignment pattern, making the cloudogram appear overly fuzzy. Rarely done, one should compare the gene-wise support: which gene supports which alternative? A neat function towards that end are the gene congruence measures implemented in IQTree or the topology-test values implemented in RAxML. When having phylogenomic data and limited number of tips, PhyloNet is the software to use. Recently, the RAxML people have introduced NetRAX, a network inference programmes. For large datasets, the simple D-statistics and ABBA/BABA test may provided crucial insights as well.
- Dissect the 35S rDNA cistron. Being a multi-copy, sometime multi-loci gene region, any in-depth analysis of this bit will give insights no other known data set can provide. The unique tandem-repeat structure may lead to ambiguous base calls because variants of different evolutionary sources are present. Typically, NGS methods will filter out intra-genomic length-polymorphism. By investigating (i.e. looking at!) the 35S rDNA sequences, which in the case of e.g. genome skimming are individual consensus sequences, one can find potentially interesting patterns for future research focusses. I happened to review such a data set quite recently. The authors lacked the experience but by just tabulating the data, I could dig out a wealth of useful information for their study.
- Make haplotype networks for the most-discriminative bits of the complete plastomes. Many apply sliding window analysis to identify the mutation hotspots in complete plastomes but few explore the signal in the identified hotspots. This is sheer negligence: as nice as phylogenomic studies are, they are not suited to solve any question at the species level because they only include one, or in the best case, a handful of individuals of a species. At the coalface evolution, even the most variable plastid sequence regions may include implicit ancestor-descendant relationships, which cannot be handled by standard tree inference. Moreover, conflicts in geographic vs. taxonomic pattering only becomes visible/ apparent when studying as many populations of a group of species as possible. For this, we need plastid marker regions with a defined discrimination capacity.
- Recruit and compare with other independent data sets. Bioclimatic data (a classic: Hijmans et al. 2005) can be really fun to add when studying generic (or inter-generic) evolution, even when done coarsely. See e.g. Grímsson et al. 2018: palaeopalynology meeting R-scripting getting into Journal of Biogeography, most notably: without any explicit biogeographic analysis! Our next paper on oaks will include Köppen profiles (weighted Köppen signatures, Bouchal et al. 2018) as well, and maybe some mapping of bioclimatic parameters.
- Last but not least: hire a palaeobotanist! Who else can make a “rich fossil record” accessible to science?
Papers like that of Areces-Berazain et al. (2021), or Li et al. (2019) and Yu et al. (2022), published in peer-reviewed Q1-journals (journal impact factors near or ≥ 5) demonstrate that too many scientists – more critical than the authors' ignorance is the peers' negligence – don't know what they have at hand and what they should be looking for. With great wealth of data comes great responsibility, to adapt a famous quote by a long-dead oil magnate. A lot of data means also a lot of opportunities to research. And not only in the blistering light – redoing what we did 15 years ago with more ("better") data, ending up with very expectable, hence, boring, results or “novel”, since totally wrong ones – but where it becomes interesting, in the dark—what we couldn't elucidate lacking the data and resources.
Mentioned or relevant literature
Areces-Berazain F, Wang Y, Hinsinger DD, Strijk JS. 2020. Plastome comparative genomics in maples resolves the infrageneric backbone relationships. PeerJ 8:e9483.
Areces-Berazain F, Hinsinger DD, Strijk JS. 2021. Genome-wide supermatrix analyses of maples (Acer, Sapindaceae) reveal recurring inter-continental migration, mass extinction, and rapid lineage divergence. Genomics 113:681–692.
Bouchal JM, Güner TH, Denk T. 2018. Middle Miocene climate of southwestern Anatolia from multiple botanical proxies. Climates of the Past 14:1427–1440.
Boulter MC, Benfield JN, Fisher HC, Gee DA, Lhotak M. 1996. The evolution and global migration of the Aceraceae. Philosophical Transactions of the Royal Society of London, Series B 351:589–603.
Cardoni S, Piredda R, Denk T, ..., Simeone MC. 2021. 5S-IGS rDNA in wind-pollinated trees (Fagus L.) encapsulates 55 million years of reticulate evolution and hybrid origins of modern species. The Plant Journal doi:10.1101/2021.1102.1126.433057.
Chen Y, Yang Q, Zhu G. 2003. Acer yangbiense (Aceraceae), a new species from Yunnan, China. Novon 13:296–299.
Delsuc F, Brinkmann H, Philippe H. 2005. Phylogenomics and the reconstruction of the tree of live. Nature Reviews Genetics 6:361-375. [Access via HAL-archives]
Denk T, Grimm GW. 2009. The biogeographic history of beech trees. Review of Palaeobotany and Palynology 158:83–100.
Denk T, Grímsson F, Zetter R, Símonarson LA. 2011. Late Cainozoic Floras of Iceland: 15 Million Years of Vegetation and Climate History in the Northern North Atlantic. Heidelberg, New York: Springer.
Denk T, Grimm GW, Grímsson F, Zetter R. 2013. Evidence from "Köppen signatures" of fossil plant assemblages for effective heat transport of Gulf Stream to subarctic North Atlantic during Miocene cooling. Biogeosciences 10:7927–7942.
Grimm GW, Denk T, Hemleben V. 2007. Evolutionary history and systematic of Acer section Acer - a case study of low-level phylogenetics. Plant Systematics and Evolution 267:215–253.
Grímsson F, Denk T. 2007. Floristic turnover in Iceland from 15 to 6 Ma – extracting biogeographical signals from fossil floral assemblages. Journal of Biogeography 34:1490–1504.
Grímsson F, Grimm GW, Zetter R, Denk T. 2016. Cretaceous and Paleogene Fagaceae from North America and Greenland: evidence for a Late Cretaceous split between Fagus and the remaining Fagaceae. Acta Palaeobotanica 56:247–305.
Grímsson F, Grimm GW, Potts AJ, Zetter R, Renner SS. 2018. A Winteraceae pollen tetrad from the early Paleocene of western Greenland, and the fossil record of Winteraceae in Laurasia and Gondwana. Journal of Biogeography 45:567–581. [Bronze OA]
Grímsson F, Bouchal JM, Xafis A, Zetter R. 2020. Combined LM and SEM study of the middle Miocene (Sarmatian) palynoflora from the Lavanttal Basin, Austria: Part V. Magnoliophyta 3 – Myrtales to Ericales. Grana 59:127–193.—Tip: All Lavanttal papers have comprehensive reference lists, ideal for lazy neotonlogists to not miss crucial palaeontological literature. And the Lavanttal Flora covers pretty much every modern plant lineage that has a fossil record, pretty much a Miocene Golden Triangle-like flora.
Heer O. 1868–1883. Flora Fossilis Arctica: Kongliga Vetenskaps Akademiens Handlingar, Stockholm.
Heřmanová Z, Kvaček J, Friis EM. 2011. Budvaricarpus serialis Knobloch & Mai, an unusual new member of the Normapolles complex from the Late Cretaceous of the Czech Republic. International Journal of Plant Sciences 172:285–293.—ususual, because it looks like modern-day Rhoiptelea but that might have triggered severe reviewer resistance.
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25:1965–1978.
Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23:254–267.
de Jong PC. 1994. Taxonomy and reproductive biology of maples. In: van Gelderen DM, de Jong PC, and Oterdoom HJ, eds. Maples of the World. Portland, OR: Timber Press, 69–104.
de Jong PC. 2002. Worldwide maple diversity. In: Wiegrefe SJ, Angus H, Otis D, and Gregorey P, editors. International Maple Symposion 02. Westonbirt Arboretum and the Royal Agricultural College in Gloucestershire, England: The National Arboretum Westonbirt. p 2–11.
Li J, Stukel M, Bussies P, Skinner K, Lemmon AR, Moriarty Lemmon E, Brown K, Bekmetjev A, Swenson NG. 2019. Maple phylogeny and biogeography inferred from phylogenomic data. Journal of Systematics and Evolution 57:594–606.
Manchester SR, Dillhoff RM. 2005. Fagus (Fagaceae) fruits, foliage, and pollen from the Middle Eocene of Pacific northwestern North America. Canadian Journal of Botany-Revue Canadienne De Botanique 82:1509–1517.
Ree RH, Moore BR, Webb CO, Donoghue MJ. 2005. A likelihood framework for inferring evolution of geographic range on phylogenetic trees. Evolution 59:2299–2311.
Renner SS, Beenken L, Grimm GW, Kocyan A, Ricklefs RE. 2007. The evolution of dioecy, heterodichogamy, and labile sex expression in Acer. Evolution 61:2701–2719.
Renner SS, Grimm GW, Schneeweiss GM, Stuessy TF, Ricklefs RE. 2008. Rooting and dating maples (Acer) with an uncorrelated-rates molecular clock: Implications for North American/Asian disjunctions. Systematic Biology 57:795–808.
Renner SS, Grimm GW, Kapli P, Denk T. 2016. Species relationships and divergence times in beeches: New insights from the inclusion of 53 young and old fossils in a birth-death clock model. Philosophical Transactions of the Royal Society B 371:20150135.
Rubel F, Brugger K, Haslinger K, Auer I. 2016. The climate of the European Alps: Shift of very high resolution Köppen-Geiger climate zones 1800–2100. Meteorologische Zeitschrift doi:10.1127/metz/2016/0816.
Sadowski EM, Hammel JU, Denk T. 2018. Synchrotron X‐ray imaging of a dichasium cupule of Castanopsis from Eocene Baltic amber. American Journal of Botany 105:2025–2036. [PDF available via DIVA-portal]
Schliep K, Potts AJ, Morrison DA, Grimm GW. 2017. Intertwining phylogenetic trees and networks. Methods in Ecology and Evolution 8:1212–1220.
Simeone MC, Grimm GW, Papini A, ..., Denk T. 2016. Plastome data reveal multiple geographic origins of Quercus Group Ilex. PeerJ 4:e1897.
Tanai T. 1983. Revisions of Tertiary Acer from East Asia. Journal of the Faculty of Science, Hokkaido University, Series IV: Geology and Mineralogy 20:291–390.
Walker JF, Walker-Hale N, Vargas OM, Larson DA, Stull GW. 2019. Characterizing gene tree conflict in plastome-inferred phylogenies. PeerJ 7:e7747.
Walther H. 1972. Studien über tertiäre Acer Mitteleuropas. Abhandlungen des Staatlichen Museums für Mineralogie und Geologie zu Dresden 19:1-309.—yes, 300+ pages, just about central European maples. Don't tell me, they are impossible to assign or irrelevant for the biogeographic history of the genus.
Wang W, Chen S, Zhang X. 2020. Complete plastomes of 17 species of maples (Sapindaceae: Acer): comparative analyses and phylogenomic implications. Plant Systematics and Evolution 306:61 [e-pub].
Wolfe JA, Tanai T. 1987. Systematics, phylogeny, and distribution of Acer in the Cenozoic of western North America. Journal of the Faculty of Science, Hokkaido University, Series IV: Geology and Mineralogy 22:1–246.—Covering more time and space but 50 pp. less than Walther (1972). Unfortunately, their morphological matrix is lost to science.
Yu T, Gao J, Liao P-C, Li J-Q, Ma W-B. 2022. Insights into comparative analyses and phylogenomic implications of Acer (Sapindaceae) inferred from complete chloroplast genomes. Frontiers in Genetics 12:791628.
Zachos JC, Pagani M, Sloan L, Thomas E, Billups K. 2001. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292:686–693.
Zhu Z, Song Z. 1999. Scientific Survey of Shennongjia Nature Reserve. ??? [in Chinese script]: ??? [in Chinese script].—We processed their lists (not in Chinese script) for Grimm & Denk (2012), Reliability and resolution of the coexistence approach — A revalidation using modern-day data, Rev. Palaeobot. Palynol. 172:33–47 (see supplement to the paper).





















No comments:
Post a Comment
Enter your comment ...