One thing about phylogenomic studies in the Era of Big Data is that we easily tend to overlook what is new and interesting. Why it may be worth to give each subtree in a phylogenomic tree a closer look
In 2016, we published an open-access paper (Simeone et al. 2016) on the western Eurasian Ilex oaks (Quercus subgenus Cerris section Ilex; Denk et al. 2017) using a few standard plastid markers. The most puzzling result was not the taxonomic decoupling or the fact that one could find several haplotypes in a single widespread species such as Q. ilex, the ‘holm oak’ [Wikipedia/Oaks of the World]. While interesting enough and worth a paper (actually two: Vitelli et al. 2017), the most puzzling was that one of the main haplotype lineages, the most diverse one, was astonishingly different from the others. We called it the ‘Euro-Med chlorotype’.
As also seen in the according ML tree, ‘Euro-Med’ was not only distinct from the other two, but likely represented an earlier derived lineage. An ancient vicariance event.
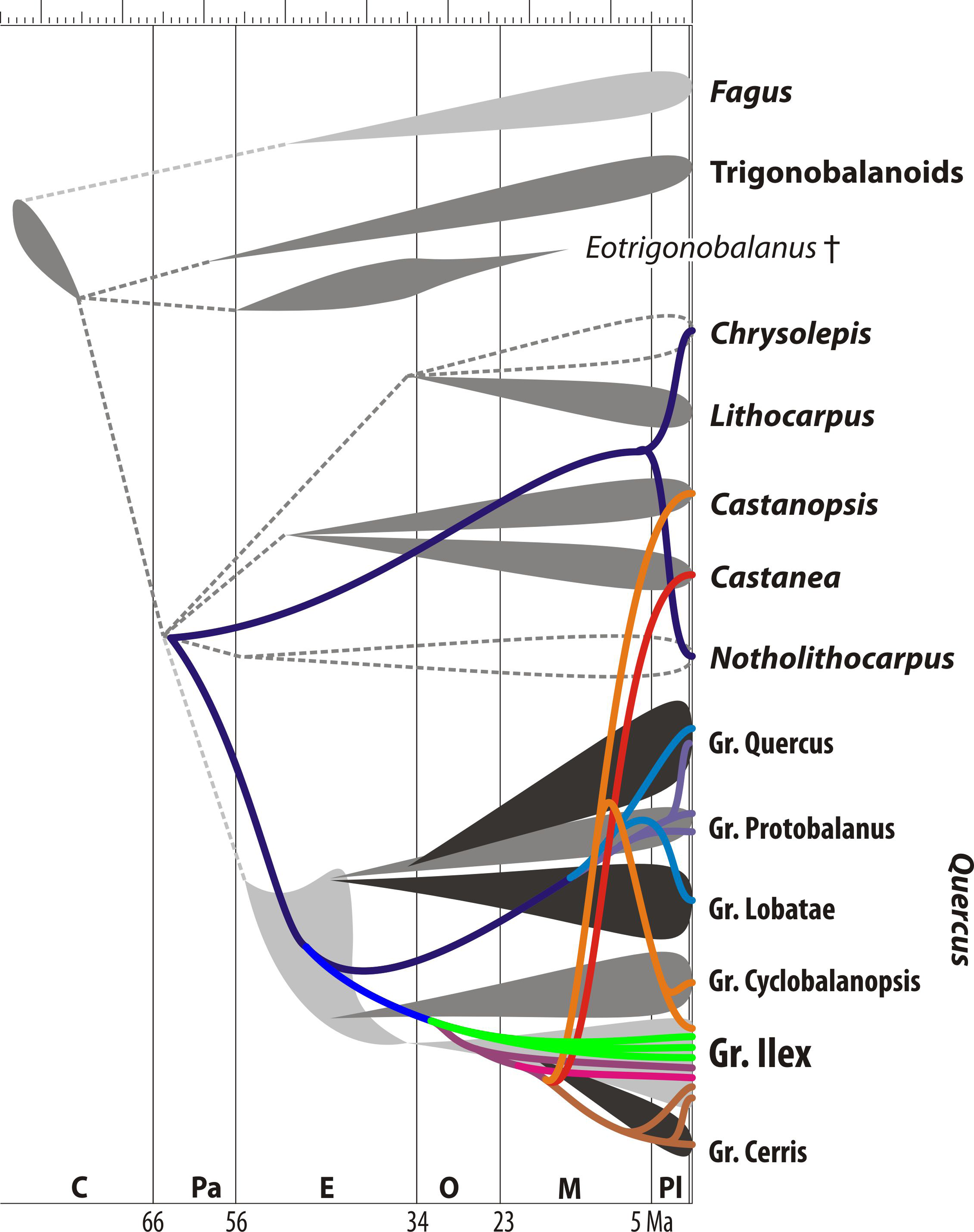
Included in the discussion of the 2016 paper was this doodle, placing these western Eurasian plastid haplotypes into a larger context. Shown in grey shades it the evolution of the family, a synoptical cactus-metaphor based on various Fagaceae studies and with respect to the fossil record (cf. Grímsson et al. 2016; green open access).
The various chlorotype lineages are coloured: Blues—the original New World (Americas, mostly N. America) core Fagaceae lineages, in reddish tones—their Old World (Eurasian) counterparts. It's a story of incomplete lineage sorting of widespread and plastid-wise already heterogenous ancestors and a few later pick-ups. What we colloquially address as “chloroplast capture”.
Glossary Chloroplast Capture and monophyly—Usually when we write this, no actual chloroplast is captured. What happens is that via (asymmetrical) introgression, one, the invading/ introgressing species takes over the population of another, the introgressed species: the F1-Hybrids and subsequent backcrosses, or introgrades in general, keep their (grand-)mother's (local) plastomes, but their nucleome is quickly homogenised by the invading species. Already the first generation may look like the fathers, the paternal donor species, when the mother's, the maternal donor species', nucleome is silenced and not expressed, but they carry and pass on an alien, to their (visible) species, plastome: the one inherited from their mother(s). Cladistically, both involved species then lost their monophyly (holophyly): the introgressing partner has one (or several) exclusively shared common ancestor(s) of the species' ancestor(s) as well as individuals and populations (or species later on) that share one with the introgressed species; the introgressed species only includes some but not all of the offspring of its last common ancestor. If the introgressed species vanishes completely, the introgressing species is technically holophyletic again, including all (surviving) offspring of the two common ancestors. When surviving the introgessed species/ lineage becomes holophyletic again as soon as all evidence for its initial (pre-introgression) radiation has been sorted out.
The phylogenomic era
Studies like ours are today often waved away as being neglectable from an evolutionary point of view, since based only on a few “fragments” and “too few” mutations (in our case, very well studied and investigated ones)—in editor-/peer-speak: “...of too narrow interest...”. The promise of phylogenomics is to get better trees (to get into higher impact journals): more basepairs = more discriminative signal = higher supported branches (not “nodes”). And, they indeed do excel when we look at the number of unambiguously supported branches. Plus: phylogenomic trees are usually results-wise trivial, we (can) take the inferred tree as the ‘true tree’ and it leaves little room for interpretation.
Or discussion. In face of a fully resolved tree with unambiguous support, we never ponder the possibility of ‘false positives’: inferred clades that are not holophyletic at all but data-/signal-/method-induced branching artefacts (All fully resolved, and perfectly misleading, species tree; but see Yang et al. 2021, for a first step in the right direction).
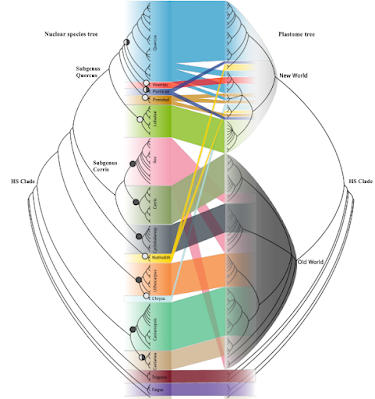
For Fagaceae, and with respect to the long known nuclear-plastid deep incongruence, we now have such a data set with a nice and comprehensive set of tips (still rare in phylogenomic studies), and for each individual up to 2000+ nuclear gene regions and their complete plastomes (Zhou et al. 2022; platin OA). A pretty unique data set allowing to infer the two corresponding, but deeply incongruent fully resolved trees.
The paper is a nice (and long expected by Fagaceae enthusiasts) piece, although it falls a bit short regarding the consequences of the now bullet-proof documented incongruence (not to mention, the commonly applied equation: high-supported inferred clade = “monophyletic”). This shortcoming has most likely to do with the fact that many peers in botany are pretty conservative in their views (there was only one in this case, who would fully appreciate what the authors' had found, and beyond).
If you look closely at Zhou et al.'s phylogenomic tanglegram and our 2016 doodle, you can see that they are very similar in any major aspect. Some people working with oaks and other Fagaceae knew (for nearly 20 years now) what treasures one can expect to dig out, if one only manages to muster the money to generate huge chunks of data.
Let's sum up the major things we can learn from Zhou et al.'s tanglegram (including some not obvious from Zhou et al.'s fig. 2):
- The New World and the Old World members of the same evolutionary lineage, i.e. a putatively holophyletic group manifesting as a clade in the nuclear trees that collects species, species groups, and genera with strong to particular phenotypic similarity, are always placed apart from each other in the plastid trees. Thus, we know that there must have been a geographic differentiation in the primordial Fagaceae that predates the divergence and manifestation of the modern-day genera and infrageneric taxa. The same we (could) see in another, equally old, northern extratropical tree lineage: the maples (genus Acer; Scientia-ex-machina...).
- This phenomenon applies at different hierarchical levels:
- Within the same section, most strikingly visible for sect. Ponticae which includes a species from the Caucasus and one from northern California, today half-a-globe apart from each other but sharing a, pretty exclusive to them (Hipp et al. 2020, gold OA; see also McVay et al. 2017), common ancestor (much back in time): the Caucasian species, Q. pontica [Wp/OotW], falls in the western Eurasian sub-subclade of the Eurasian plastid subclade of (originally New World) section Quercus; its Californian sister, Q. sadleriana [Wp-stub/OotW] groups with its pretty distant (evolutionarily) but nearby (geographically) relatives of subgenus Quercus: sect. Protobalanus.
- Within the same genus: the Eurasian oak subgenus Cerris shares a plastid lineage with the Eurasian cousins of the oaks, the chestnuts and their sisters (Castanea + Castanopsis); but the (originally) North American oak subgenus Quercus shared an all-mother with the (western) North American (near-)monotypic Chrysolepis, the North American sister of the tropical (Malesian) Lithocarpus, literally ‘stone nuts’, and Notholithocarpus, literally the 'false stone nut'. A relict genus of California extending into southern Oregon, which the most likely sister of all oaks of the world.
- Higher up: the two East Asian trigonobalanoids, Trigonobalanus (Malesia) and Formanodendron (China, endangered) have essentially the same plastome, much different from that of the South American Colombobalanus (NW. Andes), although not being sisters, only cousins (Colombobalanus is sister of Trigonobalanus).
- There's also a strict sorting within the New World and Old World plastid lineages between western Eurasia and East Asia, and less clear but obvious enough, between western and eastern North America.
Which is hardly surprising, because this is the situation 60+ million years ago.
 |
| Some earliest fossil record of Fagaceae: a pan-hemispheric family since the Late Cretaceaous (from Surviving parsimonists: just tree-naive or tree-blindfolded?) |
Nonetheless striking. And, telling a cautionary tale about the often invoked “non-monophyly” and “out-of-East Asia/China” scenarios based on plastid or complete plastome data or blindly combined nuclear-plastid data sets (Are complete plastome trees always better?; How not to make a phylogeographic study; Oak systematics and complete plastome trees; Scientia-ex-machina: explicit biogeographic inferences and the phylogenomic age).
Or fully resolved “species trees” based on nothing more than (inevitably undersampled) single-individual (worse: cultivated of unknown provenance) complete plastome trees. And if one dives into the details of Zhou et al.'s tangling nuclear and plastid trees...well, it is just the beginning of a wonderful journey into the we-have-never-heard-of-Hennig-or-Mayr evolution of this family and its few but puzzling genera. In fact, based on these data one could have written all there is to say about Fagaceae evolution. But, that would have been too many pages and much too deep for higher flying journals like Nature Communications.
[PS Any systematicist who wants to remain a cladist should read Wheeler (2014) and get accustomed with terms like “epiphyly”, “periphyly” and “anaphyly”, you'll need them.]
The missed (and lost) lineage
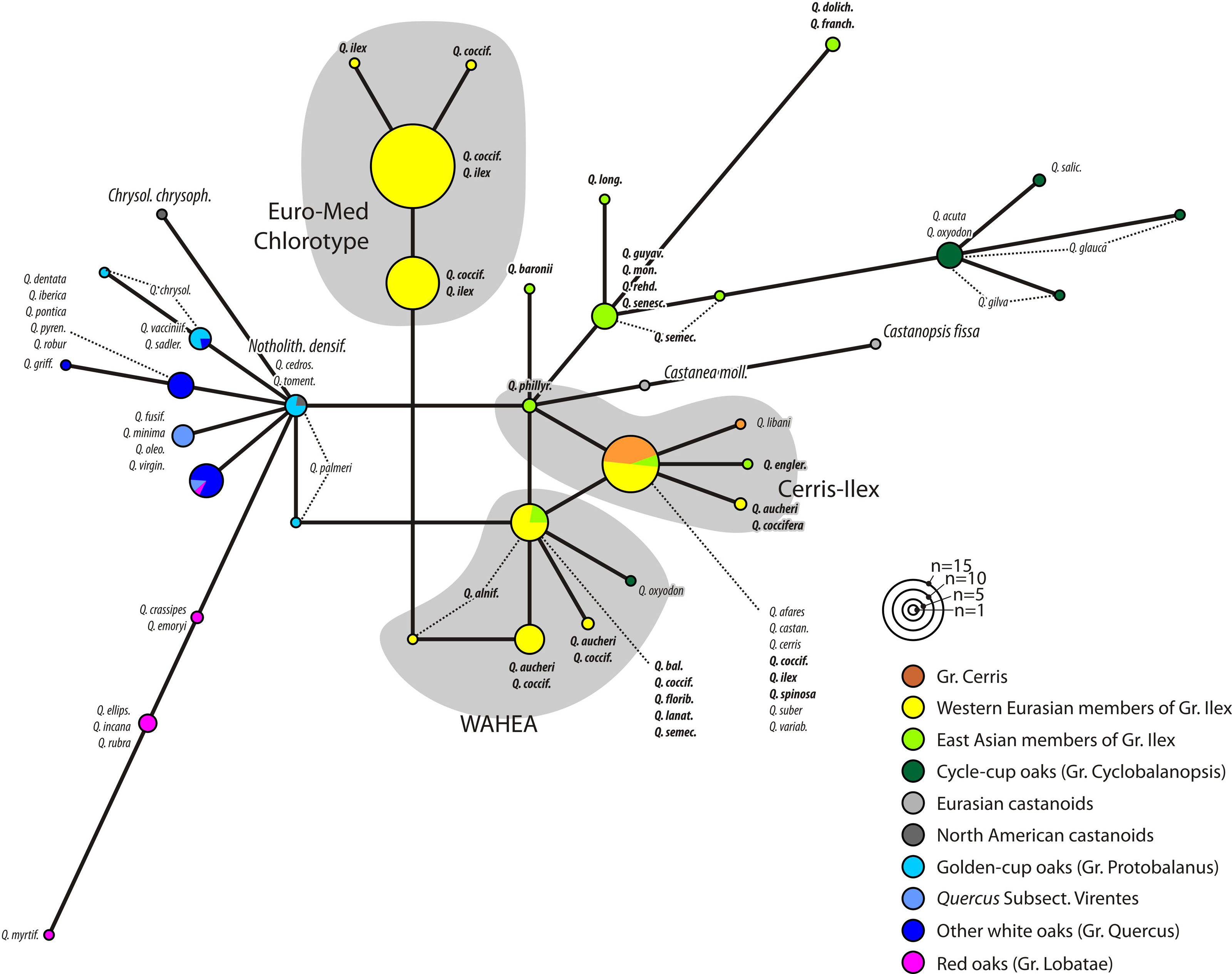
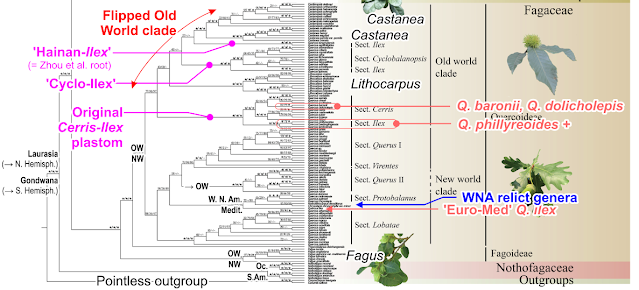
When looking at the supplement trees, noting which species has which plastid lineage or not, we can map the pink group, section Ilex, and our 2016 chlorotypes. Here's which clade/MRCA in Zhou et al.'s tanglegram corresponds to our 2016 lineages.
As nice and comprehensive as the trees are, they miss our ‘Euro-Med’ chlorotype; i.e. the most interesting of all (still living) oak plastomes. Zhou et al. (2022) didn't include any Q. ilex in their study, the most widespread of all western Eurasian Ilex oaks. From the fossil record we know that both section Cerris and Ilex likely originated in north-eastern Asia, hence their similar plastomes (uppermost pink group). Any of the other plastomes must then have been picked up by section Ilex on its way south and then west along the “Himalayan corridor”. Possibly including ancient East Asian ones, the bottom-most pink group I labelled “Genuine East Asian Ilex”. Looking at our 2016 tree, we can nonetheless make a guess where the lost lineage plastome(s) would pop up, at the very basis of the whole Old World core Fagaceae clade.
Why is the ‘Euro-Med’ the most interesting of all Fagaceae chlorotypes? Fagaceae plastomes are notoriously conserved. Looking at our 2016 graphs, a handful of mutations in the trnH-psbA intergenic spacer, adding to not more in the other marker; it doesn't look like much of a differentiation. But keep in mind:
- The intra-clade differentiation in the ‘Euro-Med’ plastid lineage outcompetes what we see between North American and Eurasian species of section Quercus (which probably have diverged later but not recently) and between East Asian and western Eurasian species of sections Cerris and Ilex sharing the ‘Cerris-Ilex’ s.l. plastomes (including the ‘WAHEA’ and further, similar East Asian chlorotypes). Based on our latest dating (using nuclear-SNP data and a shitload of fossils to inform the clades' age ranges; Denk et al., submitted: pre-release data dump and supplement available via Zenodo), we can place this radiation in at least early Oligocene times (related tweet-thread; chronogram shown below). Same seems to hold for the other plastid lineages we find in East Asian (into the eastern Himalayas) Ilex oaks.
- The inter-clade differentiation in the same marker is higher between the ‘Euro-Med’ haplotypes and the partly overlapping, also Mediterranean ‘Cerris-Ilex’ (s.str., i.e. western Eurasian Cerris-plastomes picked up by western Eurasian Ilex oaks) and ‘WAHEA’ haplotypes (shared by Euro-Mediterranean and W. Asian Ilex oaks) than between the latter two; or between them and the other ±related East Asian Ilex oaks. Species whose individuals carry plastomes (the ‘genuine E. Asian Ilex’ in the figure above) that partly diverged before even those of the chestnuts and their tropical-subtropical sisters, Castanopsis, genera with a fossil record going back into the Eocene (>45 Ma).
- In the same markers, even less mutations differ between the oaks and other Fagaceae genera of the same continental lineage.
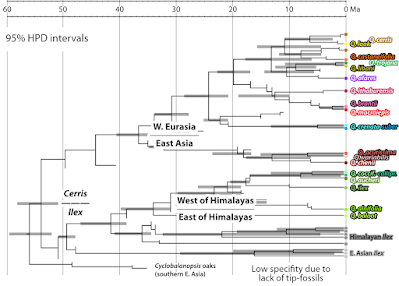 |
| Fossilised-Birth-Death modelled chronogram of subgenus Cerris (modified after Denk et al., submitted, Supplementary fig. 4). Grey bars: 95% HPD confidence intervals. |
We knew for some time and have now irrefutable proof thanks to Zhou et al. (2022) that the Ilex oaks, the sister lineage (in a strict evolutionary sense) of sect. Cerris, the cork oaks (see also Hipp et al. 2020), love(d) picking up alien plastomes: “chloroplast capture”. They most probably did it (and some may still be still doing it) by (massive) asymmetric introgression: an Ilex newcomer migrates into another species' (cousin's) area, with the hybrids/ introgrades crossing back with or being pollinated mainly by the Ilex invader, until the native species has been taken over. Intra-species homogenisation wipes out any nuclear evidence of the other parent species but the taken-over native mother trees propagate their chloroplast plasmids into their introgressed daughters, grand-daughters and so on. Over millions of years till today in the absence of severe bottlenecks leading to the extinction of the now alien to the species mothers.
From Zhou et al.'s tanglegram and our 2016 few-bits study, we can deduce that the Ilex oaks did this three or four times at a large(r) scale: when going south, they took over cycle-cup oaks (sect. Cyclobalanopsis, the sister lineage of Cerris-Ilex) forming section Ilex species with Cyclobalanopsis plastomes. They also picked up an even earlier diverged plastome, the “Genuine East Asian Ilex”, if section Ilex was originally holophyletic (as indicated by past and modern morphology and the nuclear data). After going west via the Himalayan corridor, the Ilex oaks came in secondary contact with their further evolved section Cerris sisters (who took a different path when leaving north-eastern Asia; Denk et al., submitted) and exchanged locally their plastids. Notably there's no sign so far of (evolved, unique) Ilex-plastomes in East Asian Cerris oaks or vice versa, but we do have western Eurasian Cerris-plastomes in Mediterranean Ilex and v.v. (Vitelli et al. 2017; Simeone et al. 2018, gold OA). And when they arrived in the (western) Mediterranean (in the Oligocene), they could have encountered an ancient, aboriginal European oak species, hybridising with it, introgressing it, eventually forming one of the precursors of today's Q. ilex, some of which still carry that lost lineage's plastome.
The overlooked tip
I'm out-of-business, and the people I worked with and for were always notoriously short on resources to make a big splash, molecularly. While we could have easily got our hands on material, it soon became impossible to get the money needed to do up-to-date, massive-data molecular analysis, not to mention the money to hire the hands needed to sequence and process complete ‘Euro-Med’ plastomes. When I checked a few days ago, gene banks had 199 complete plastome accessions (including duplicates of NC… reference sequences) and still no Q. ilex, the name-giving and most important of all Ilex oaks. But, luck striking, such a plastome may have been accidently, and unknowingly, already been produced.
Completely unrelated to my old passion for beeches and oaks, I was recently pointed to another quite fresh-from-the-press paper by Yang et al. (2021) showing an all-Fagales complete plastome tree.
This tree is remarkable because it confirms that all classic plastid “barcode” trees have been pretty wrong (outside the Fagaceae) regarding many data- and signal-wise non-trivial relationships, which includes the phylogenies relied on in much-cited journals and papers: the original “three genome” tree of Li et al. (2004; see All solved a decade ago: the asterisk branch in the Fagales phylogeny) – until today the basis for Fagales phylogenetics and systematics – and the artficial ITS-plastid topology relied on by Sauquet et al. (2012)—published in the phylogeny flagship journal Systematic Biology. Not to mention the even worse data set and tree used by Larson-Johnson (2016); published in the equally reputable journal New Phytologist. Well, reviewers simply lack the time to look into the data used for fancy analyses.
While complete plastome trees do not necessarily outperform dense-sampled, low-level trnH-psbA or alike plastid bits data (Are complete plastome trees always better?), they can outperform the classic (typically matK-constrained; Why the emperor has new clothes on – the mighty matK) oligo-(“multi-”)gene plastid trees. For example, all plastid-based or -constrained earlier trees except ours from 2013 found Alnus, the alder, sister to the remaining Betulaceae. But Yang et al., using the concatenated plastome data, recovered exactly our 2013 topology, with Alnus sister to birch, Betula. Why did we get a different topology, one only confirmed by Yang et al.'s (near-)complete plastome data eight years later? Because for our Betulaceae paper, we used all plastid data we could possibly harvest from gene banks, while all Fagales studies before and after us kept themselves to the old (and in this case partially misleading: All solved a decade ago...) classics of Li et al. (2004). Trying something new and as open-minded as in-depth (Grimm & Renner 2013) won't get you in the best-of-journals, but you'll may get a more sensible tree a decade ahead of everyone else :D
Yang et al.'s tree is furthermore remarkable because it shows in one picture the two completely different sorting modes in the Fagales (when knowing the phylogenetic, nuclear-data-based relationships): the multi-level plastid decoupling in the Fagaceae, the 2nd divering Fagales lineage vs. a much higher nuclear-plastid congruence above and at the genus level in the core Fagales (Betulaceae, Myricaceae, Juglandaceae). Naturally, whenever a family or genus covers New and Old World species, it gets split up and deviates from the rare (outside Fagaceae) nuclear-based trees generated so far. Except for alders and birches; but they can disperse long-distance (literally, over hundreds of kms) their seeds as easily as their pollen (and another exception, but that's a topic for another post). In the case of the 1st diverging and only entirely Gondwanan, today southern hemispheric Fagales lineage, the Nothofagaceae, it's Oceania vs S. America; at the inter-(sub-)generic level contrasting the nuclear data as well.
The oak sections are annotated in Yang et al.'s tree, we can see “sect. Ilex” pops up three times like in Zhou et al.'s plastome cladogram.
- One East Asian species is placed as sister to Castanea-Castanopsis plastomes, which I tentatively labelled the “Hainan-Ilex” plastome (see below); this is the same chlorotype than the first diverging plastid lineage in Zhou et al.'s Old World clade (‘genuine E. Asian Ilex lineage’ above).
- A couple of species are placed as sister to Quercus sect. Cyclobalanopsis plastomes, these are the Ilex oaks that introgressed their tropical-subtropical sisters when going south, let's call them “Cyclo-Ilex”.
- as sister to section Cerris: “Cerris-Ilex” in the most comprehensive sense, i.e. including both our 2016f “Cerris-Ilex” and “WAHEA” chlorotypes as well as further East Asian Ilex plastomes that form part of the same plastid lineage.
Non untypical for large-scale analysis is the floppiness of the annotation (Quercus missing a r, to my own experience, very few reviewers look at pictures no matter how much work you put into them) or bracketing to the right: Q. baronii and Q. dolicholepis, to central East Asian Ilex oaks, come as sister to the East Asian members of section Cerris and splitting them from their western Eurasian sisters (which may be an alignment-data branching artefact related to using unchecked plastome data; terminal subtrees close to the speciation level are notoriously difficult to resolve in Fagales). The authors just included them in the sect. Cerris bracket. Either by accident or to not draw any attention to the misfit.
Another notable difference is that the entire Old World Fagaceae subtree is flipped: while in Zhou et al. (2022), the ‘Hainan-Ilex’ plastomes diverge first, followed by Castanea-Castanopsis, they now form the core group. Or, cladistically speaking: they change from a assumedly paraphyletic “basal grade“ into a holophyletic per definition “crown clade“. Another fine example why we really should stop synonymising inferred (molecular) clades with “monophyletic” (never a good idea for plastid clades, and not necessarily for nuclear ones as well).
The tip we are interested in, is one not bracketed by any of the oak labels to the right. It's the one labelled as a Q. ilex. It's in the wrong clade! An Old World plastome, the type species of sect. Ilex, embedded in the New World Fagaceae.
Now, mislabellings are pretty common, and increasingly so in the age of Big Data. But this one pops up, where you well would picture the plastome of a long lost ancient and native European lineage: as sister to a clade comprising the (morphologically and genetically) least evolved, but early diverged and exclusively New World core Fagaceae: the (quasi-)monotypic western North American genera Chrysolepis and Notholithocarpus and sect. Protobalanus of subgenus Quercus. The former two are impossible to discern in the fossil record, but the latter has been exclusively a New World lineage already in the early days of oaks (Bouchal et al. 2016). This Q. ilex may have what no other Q. ilex individual used outside our own work has had so far: a genuine ‘Euro-Med’ plastome, captured from a long-gone European but not really “Old World” oak.
What a single plastome may tell us about the early evolution of Fagaceae (and Fagales)
Odd, isn't it? This cladogram indicates with no ambiguity (the first value gives the ML branch support based on the concatenated matrix) that the ‘Euro-Med’ type defies the general rule within Fagaceae, and most Fagales—a western Eurasian species has always an Old World and never a New World plastome. In addition, this putative ‘Euro-Med’ plastome is part of a clade comprising only western North American relicts, left-overs of early radiations from the other side of the world (for sect. Protobalanus, we know it occurred also east of the Rockies main chain at the Eocene-Oligocene boundary; Bouchal et al. 2016). Why not at least eastern North American, the region that has been connected for a long time directly to Europe via the North Atlantic Land Bridge?
Because it's not just a plastome from some cross-Atlantic dispersed North American white (sect. Quercus) or red oak (sect. Lobatae), but a plastome picked up from a very ancient oak lineage. A possibly third, extinct subgenus that formed before the two modern ones, the New World subgenus Quercus and the Old World subgenus Cerris, radiated.
Picked up, when ~10-15 myrs later the first common ancestor(s) of the modern-day western Eurasian Ilex oaks invaded what would become the Mediterranean and came into contact with the leftovers of the once widespread across Europe but also North America “paratropical” or “boreotropical” forests; and the primordial oaks thriving in them, then still relatively close relatives.
If Yang et al.'s tree is not completely biased, the ‘Euro-Med’ type is the legacy of an ancient cross-Atlantic oak lineage; a lineage that was established not long after the first oaks evolved and radiated.
We don't know for sure (yet; would need a joint palaeo+neobotanical effort). What we know is that these (possibly) even more ancient (American-)European sisters or cousins left no survivors. But the genetic-palaeogeographic legacy of the lost Palaeogene (Atlantic-)European lineage lives on in the western Mediterranean populations of modern-day Q. ilex: the ‘Euro-Med’ chlorotype.
It may be just a single, unbracketed, not annotated tip in a large-scale complete plastome tree, but a most interesting one.
Demonstrating how important (and valuable) the now a bit frowned upon classic “fragment”-studies (few markers, lots of individuals) still are. In particular, when you want to pick the best and most comprehensive sample for sequencing complete plastomes towards increasingly trivial and easy-to-infer but nonetheless still tricky-to-interpret trees.
Cited literature
Green font—crucial introductory [quasi-]open access reads for anyone interested in Fagaceae/oaks, including peers reviewing papers on them; grey font—pointless read, irrespective of interest; being fundamentally flawed (poor data control).
- Bouchal JM, Zetter R, Denk T. 2016. Pollen and spores of the uppermost Eocene Florissant Formation, Colorado: a combined light and scanning electron microscopy study. Grana 55:179–245.
- Denk T, Grimm GW, Manos PS, Deng M, Hipp AL. 2017. An updated infrageneric classification of the oaks: review of previous taxonomic schemes and synthesis of evolutionary patterns. In: Gil-Pelegrín E, Peguero-Pina JJ, and Sancho-Knapik D, eds. Oaks Physiological Ecology. Cham: Springer, 13–38. Free Pre-Print at https://doi.org/10.1101/168146 [major change: Ponticae and Virentes accepted as additional sections in final version]—this paper represents the currently valid oak systematics.
- Grimm GW, Renner SS. 2013. Harvesting GenBank for a Betulaceae supermatrix, and a new chronogram for the family. Botanical Journal of the Linnean Society 172:465–477. [PDF]
- Grímsson F, Zetter R, Grimm GW, Krarup Pedersen G, Pedersen AK, Denk T. 2015. Fagaceae pollen from the early Cenozoic of West Greenland: revisiting Engler's and Chaney's Arcto-Tertiary hypotheses. Plant Systematics and Evolution 301:809–832.
- Grímsson F, Grimm GW, Zetter R, Denk T. 2016. Cretaceous and Paleogene Fagaceae from North America and Greenland: evidence for a Late Cretaceous split between Fagus and the remaining Fagaceae. Acta Palaeobotanica 56: 247–305.
- Hipp AL, Manos PS, Hahn M, Avishai M, ...[a few others in alphabetical order]...Valencia Avalos S. 2020. Genomic landscape of the global oak phylogeny. New Phytologist 226:1198–1212.
- Hubert F, Grimm GW, Jousselin E, Berry V, Franc A, Kremer A. 2014. Multiple nuclear genes stabilize the phylogenetic backbone of the genus Quercus. Systematics and Biodiversity 12:405–423.
- Larson-Johnson K. 2016. Phylogenetic investigation of the complex evolutionary history of dispersal mode and diversification rates across living and fossil Fagales. New Phytologist 209:418–435.
- Li R-Q, Chen Z-D, Lu A-M, Soltis DE, Soltis PS, Manos PS. 2004. Phylogenetic relationships in Fagales based on DNA sequences from three genomes [in reality: four plastid “barcodes”; the only nuclear gene, 18S rDNA, is highly incongruent and outcompeted, and the only mitochondrial gene, has no discriminative capacity at all]. International Journal of Plant Sciences 165:311–324.
- McVay JD, Hipp AL, Manos PS. 2017. A genetic legacy of introgression confounds phylogeny and biogeography in oaks. Proceedings of the Royal Society B 284:20170300.
- Sauquet H, Ho SY, Gandolfo MA, Jordan GJ, Wilf P, Cantrill DJ, Bayly MJ, ... Udovicic F. 2012. Testing the impact of calibration on molecular divergence times using a fossil-rich group: the case of Nothofagus (Fagales). Systematic Biology 61:289–313.
- Scotese CR. 2013. Oligocene Globe (Oligocene_Pgeog_850.kmz, Google Earth format), www.globalgeology.com, PALEOMAP Project, Evanston, IL. ResearchGate data set.
- Scotese CR, Boucot AJ, Xu C. 2014. Atlas of Phanerozoic climatic zones (Mollweide Projection), vols 1–6. PALEOMAP Project PaleoAtlas for ArcGIS. Evanston, IL.: PALEOMAP Project.
- Simeone MC, Grimm GW, Papini A, Vessella F, Cardoni S, Tordoni E, Piredda R, Franc A, Denk T. 2016. Plastome data reveal multiple geographic origins of Quercus Group Ilex. PeerJ 4:e1897.
- Simeone MC, Cardoni S, Piredda R, Imperatori F, Avishai M, Grimm GW, Denk T. 2018. Comparative systematics and phylogeography of Quercus Section Cerris in western Eurasia: inferences from plastid and nuclear DNA variation. PeerJ 6:e5793.
- Vitelli M, Vessella F, Cardoni S, Pollegioni P, Denk T, Grimm GW, Simeone MC. 2017. Phylogeographic structuring of plastome diversity in Mediterranean oaks (Quercus Group Ilex, Fagaceae). Tree Genetics and Genomes 13:3.
- Wheeler WC. 2014. Phyletic groups on networks. Cladistics 40:447–451.—see also this post by D. Morrison on Genealogical World of Phylogenetic Networks
- Zhou B-F, Yuan S, Crowl A, Liang Y-Y, Shi Y, Chen X-Y, An Q-Q, Kang M, Manos P, Wang B. 2022. Phylogenomic analyses highlight innovation and introgression in the continental radiations of Fagaceae across the Northern Hemisphere. Nature Communications 13:1320.





No comments:
Post a Comment
Enter your comment ...