The ultimate promise of phylogenomics is to get a fully resolved species tree: a tree, where the individuals finely sort per species, and where all branches, especially the deepest ones, have high or, better, unambiguous support. A look behind such a tree, Jiang et al.'s (2021) Tree of Beeches.
Beech trees were my first study foes, together with maples. Back at the beginning of the new millenia, we (the late Karin Stögerer, our technician in the web lab, and I, mainly on the computer) generated over the years hundreds of ITS sequences of beeches and maples. We quickly realised that the promises of much-cited papers such as Álvarez & Wendel (2003, Mol. Phylogenet. Evol. 29, 417–434) didn't apply to temperate trees. Indeed, our cloned ITS data provided quite some insights into the phylogeny of these genera. But they – in general and the closer one came to the tips of the Tree of Life – clashed with the concept behind any phylogenetic tree: that speciation is a dichotomous process, and time the only dimension. The ITS data pointed to occassional (in case of maples) and common (in case of beech) reticulation, inter-species gene flow and intra-species diversity, population dynamics messing up the ITS pools' phylogenetic signals.
Two decades later, Big Data have replaced our ITS data, and phylogenomic data sets have provided ... well, in the case of maples, nothing fundamentally new really (Are complete plastome trees always better...) More maple species have been covered, a few intriguing ones, and others much too poorly. Using data from a single arboretum specimen for known heterogenous species (including a few that may be misidentified, or cultivation hybrids; e.g. accessions of Acer sect. Acer used by Areces-Berazain et al., 2020, PeerJ 8:e9483) may not be the best path to follow. Especially, when one objective is to get a better species tree. But, the support for the deep and some critical branches has increased.
While the big evolutionary questions posed by early ITS and "plastid DNA fragment" data on Acer are still unanswered entirely, for the about a dozen species of beech, we now have a fine, fully resolved species tree.
Jiang L, Bao Q, He W, Fan D-M, Cheng S-M, López-Pujol J, Chung MG, Sakaguchi S, Sánchez-González A, Gedik A, Li D-Z, Kou Y-X, Zhang Z-Y. 2021. Phylogeny and biogeography of Fagus (Fagaceae) based on 28 nuclear single/low-copy loci. Journal of Systematics and Evolution doi:10.1111/jse.12695.
Most importantly, a species tree including more than one individual per species and only individuals collected from the wild.
A perfectly resolved tree
Jiang et al. (copied from abstract)
... sequenced 28 nuclear single/low‐copy loci (18 555 bp in total) of 11 Fagus species/
segregates and seven outgroups. Phylogenetic trees were reconstructed using both concatenation‐based (maximum parsimony, maximum likelihood, and Bayesian inference) and coalescent‐based methods (StarBEAST2, ASTRAL).
and found
The monophyly of two subgenera (Fagus and Engleriana) and most [monotypic, i.e. including only a single species] sections was well supported, except for sect. Lucida [two species], which was paraphyletic with respect to [monotypic] sect. Longipetiolata. We also found a major phylogenetic conflict among North American, East Asian, and West Eurasian lineages of subgen. Fagus. Three segregates that have isolated distribution (F. mexicana, F. multinervis, and F. orientalis) were independent evolutionary units.
Reziprocal monophyly of Shen's (still informal, hence, not be to italicised) subgenera is a phylogenetic scenario obvious from ITS data only. Hence, we used it in 2016 when dating inter-specific divergences (Renner et al., 2016).
The uniqueness of F. multinervis can also be seen in LEAFY intron data generated by a Korean study in 2016 (Oh et al., 2016; see also Worth et al., 2021).
For the complex genetics of the western Eurasian beeches, F. sylvatica (s.str.) vs. F. orientalis, check out this paper:
Gömöry D, Paule L. 2010. Reticulate evolution patterns in western-Eurasian beeches. Botanica Helvetica 120:63–74.
Documenting the genetic distinctness of F. mexicana from F. grandifolia is a nice finding, although hardly surprising given 1500 km air-distance, and the need to cross either open sea or semi-desertic steppes to get from the southernmost F. grandifolia (s.str.) in northernmost Florida to the F. mexicana populations in central Mexico.
The tree also shows that the continental (PRC) and insular (Taiwan, i.e. ROC) populations of F. hayatae are equally distinct. I suppose this would have been politically hazardous to highlight for Chinese authors funded by the state and publishing in a Chinese journal. The authors just collapsed the according subtree for their main-text fig. 1 and don't mention this "segregate" at all (which is a pity, because that's actually a nice and very interesting find as we will see).
Overall, the beech phylogenomic data provide some new insights and the new species tree successfully settled some disputes. And there can be few follow-up questions, looking at the resolution of the species tree.
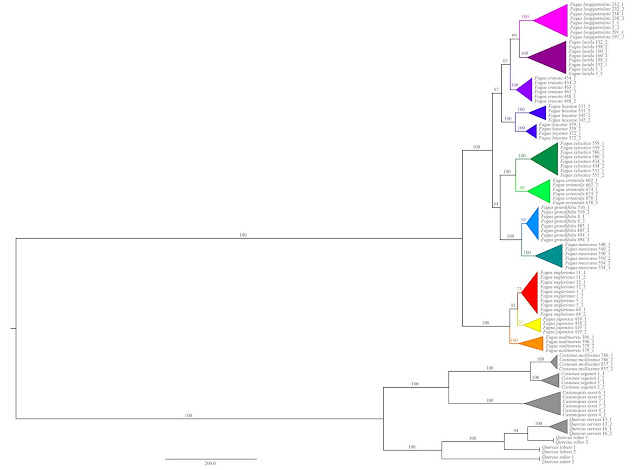
| Jiang et al.'s outgroup-rooted species tree (as provided in Supplement file S1). All species-level taxa are finely sorted and unambiguously supported, same holds for the backbone, the inter-species relationships. Note the extreme divergence between Fagus and the other Fagaceae, a phenomen observed in any genetic data generated so far, inevitably leading to ingroup-outgroup branch attraction: the root is placed at the most distinct ingroup split, which is the subgeneric one. The "1" and "2" indicate the phased haplotypes used by Jiang et al. instead of the original sequence data. |
Why I couldn't believe the tree
The simple answer is: it's a fully resolved tree based on "single-/low-copy" nuclear genes, and I know the signal in single- and multi-copy non-coding, hence faster evolving, nuclear sequences: not overly tree-like, and not in perfect agreement.
- Even if all genes follow the same phylogeny, i.e. have the same gene histories, the low-copy ones would include paralogues. Which should mess things a bit up because of incomplete lineage sorting. But, they don't.
- We know for the western Eurasian Fagaceae, beeches and oaks, that there has been a lot going on between the species. And the few Chinese Fagaceae species studied intensively, show the same (Why you should never do a single-species plastid analysis of oaks)
Jiang et al. note that
... contemporary hybridization events in the genus should be considered the exception and not the norm, both within and outside China. However, this fact indicates that the beech species or segregates recognized in this study [F. mexicana, F. multinervis, F. orientalis] might have experienced long independent evolutionary histories that have allowed them to accumulate private mutations through genetic drift and selection.
In slow-evolving, probably coding nuclear gene regions, but not in the ITS?! What could be the trigger? (Spoiler: it's reticulation, the "not the norm" stuff.) In Jiang et al.'s tree F. longipetiolata is the sister of F. lucida and F. hayatae s.l. are sister to all other East Asian species of subgenus Fagus. This is a bit different what other data sets pointed to.
Based on intra-individual ITS polymorphim, F. lucida is closest to F. crenata (Grimm et al. 2007), the forth East Asian species of subgenus Fagus (green split in the graph above). While the sympatric F. longipetiolata and F. hayatae subsp. pashanica, the continental, Chinese F. hayatae, share a conspicious ITS dimorphism (Denk et al. 2005; we didn't had access to any proliferate Taiwanese F. hayatae specimen). In addition to the Eurasian-shared ITS variants found in all Eurasian species of subgenus Fagus (ITS lineage IV in the figure below, grey), they exclusively share a "private", substantially distinct ITS type (lineage III, blue).
Jiang et al. – using wrongly placed fossils, hence, 20–30 myrs too young age priors (see also The most common errors regarding node dating) – dated the MRCA of the ITS-dimorphism sharing F. hayatae and F. longipetiolata to ~8 Ma. Convergent, directed evolution leading to high-diagnostic ITS variants, like those of Fagus lineage III, is biologically/ genetically improbable. If we take Jiang et al.'s tree, the only way the not-sister species F. hayatae and F. longipetiolata caught up their type III ITS arrays has been via a "not the norm" hybridisation event in the last 8 myrs. III could be the original F. hayatae type (first diverging lineage in Jiang et al.'s East Asian crown clade) and IV the type shared by all other East Asian Fagus species. It could also be a more ancient dimorphism, result of an ancient reticulation and/or 35S rDNA array duplication in the common ancestor of the Eurasian Fagus clade, and the other species eliminated their type III ITS variants, independently from each other (three times: ancestor of the western Eurasian beeches, of F. crenata, and of F. lucida).
The curiously conflicting data behind the fully resolved tree
Having being pointed to this just-out tree and new data during the review of an upcoming paper looking at the 5S intergenic spacer diversity in a few selected beech individuals (Cardoni et al., 2021), I got intrigued by it. There were two more things that got me curious:
- The fuzzyness of the cloudogram of Jiang et al. (2021, fig. 2), the *BEAST result. It's simply too fuzzy with respect to the support in the combined tree. Additional links included F. crenata-F. hayatae sister relationship and the alternative of an Euamerican clade, a sister relationship between the North American and western Eurasian species. Those reading palaeontological literature – not a frequent sport among phylogenomic biogeographers and daters (not Jiang et al. 2021, who took what fitted them from our 2009 paper) – know that the North Atlantic land bridge was used by beeches (Grímsson & Denk, 2005, 2007; Denk & Grimm, 2009). And we did find a North American (lineage II) pseudogenic ITS copy in one of our Caucasian (Eastern F. orientalis) individuals (Denk et al. 2002), and F. grandifolia is not cultivated in Georgia.
- The use of phased, reconstructed haplotypes rather than the original gene data. In the absence of recent or contemporary hybridisation, assumedly diploid genomes (but see Ribeiro et al., 2011) should not be dramatically heterozygotic, carrying around more than a single gene variant ending up in polymorphic base calls. But it says "single-/low-copy" in the title. So, there may be paralogues? How polymorphic were the originally obtained gene sequences, that the authors saw no other way out than to phase each gene into two haplotypes per sampled individuum?
When one looks at the phased haplotype data – an even rarer sport among phylogeneticists than keeping up with palaeontological literature – one can see that Jiang et al.'s individuals include heterozygotes and are generally rich in intra-individual sequence polymorphisms (2ISPs, Potts et al. 2014). Which is odd, given Jiang et al.'s premises. If hybridisation, inter-species gene flow in general, is not "normal", such polymorphism can only be the result of gene duplication. Which would mean most of the 28 genes are low-copy, since we find 2ISPs in every one, ranging from two in gene P54 to 36 in gene F202.
By de-phasing the data, building the strict consensus of the two documented reconstructed haplotypes per individual (i.e. the data uploaded to gene banks), I de-constructed the original individual-level genetic data.
The de-phased, putatively original sequence data enables us to explore the signal in the 2ISP patterns and individual-/species-wise sorted mutations (mostly SNPs—single-nucleotide polymorphisms; all tabulated in Supplementary file S5 to Cardoni et al., 2021). Which allows to reject pretty much anything Jiang et al. considered. In particular, the principal assumption behind all their reconstructions and interpretations: that speciation in beech is primarily a dichotomous process.
It's not, and has never been. Following different evolutionary trajectories, the individual nuclear genes assembled by Jiang et al. furbish a genetic kaleidoscope of reticulation and incomplete lineage sorting. A glorious genetic mess. Something rarely documented so far for such a fine genus sample (nearly all species, natural stands only, 28 nuclear genes), and utterly ignored by the authors themselves. And most likely, by any reader of the paper.
Hence, this post.
Emerging paralogy
All 28 genes are all low-divergent. Furthermore, there is relatively little intra-individual length-polymorphism in the sample (at least not in the documented data; there are some conspicious data gaps, though). Alignment and investigation of point mutation and 2ISP patterns is straigthforward. Changing from gene to gene, some individuals are 2ISP-rich and apparently heterozygotic/ poly-genetic, while others show pure genotypes without any 2ISPs (or only randomly distributed ones). Hence, by just tabulating the mutation-/2ISP-patterns (data link provided at end of post), we can identify the main gene variants and potential paralogues and map them on the combined tree.
In case of gene F202, we can discern four major variants (A to D), A and B exclusive to subgenus Engleriana, C and D to subgenus Fagus. The A and B variants are not sorted by species at all, the C and D to some degree. In case of variant A and C, ancestral gene variants persist in the gene pool of a single individual (F. japonica, pink lining) and all modern-day Eurasian species of subgenus Fagus (grey). While most individuals only show one (sub-)variant (filling colours fit according species' colours), of all members of subgenus Engleriana F. multinervis – the relict Engleriana of Ulleungdo and first branching Engleriana species in the combined tree – combines the A and B variants otherwise found in Chinese F. engleriana. Possibly an ancient polymorphism rather than recent gene flow given the >1600 km air-distance between the contemporary populations of both species (again, they would have to cross open water and tree-free steppes). The heterozygotic F. mexicana vs. homozygotic F. grandifolia (s.str.) demonstrate gene loss in the North American lineage. This could be linked to high levels of intra-species homogenisation: one of the two subvariants got lost in the widespread, huge population-sized north-eastern species. An alternative explanation is asymmetric introgression during the Pleistocene fluctuations when the grandifolias had to migrate south. Inter-species relationships in Eurasian Fagus are highly complex. The hayataes are distinct and share a specific general D variant, fitting with the combined tree. The only longipetiolata-lucida link, however, is the retention of the ancestral C variant; progressively replaced by a specific subvariant in F. lucida, while F. longipetiolata boosts his own D variants. The western Eurasian species and their closest (based on ITS data and morphology) living relative, the Japanese F. crenata, share both their C and D variants, in strong conflict with their position in the combined tree. The western Turkish individual (Western orientalis) and one of the southern ones (orientalis of yet to be defined affinity; cf. Gömöry & Paule 2010) have replaced the ancestral C already by their own, derived subvariant (light green).
High support masking ancient reticulation
The next picture is a visualisation of the gene variants found in the sampled individuals, colour-coded for ancestry (primitive-shared; grey, pink, yellow) and specifity (derived, unique sequence types; all other colours) and mapped on a simple unpartitioned tree (left) that can be inferred on the combined individual-level data (Grimm, 2021). The colouring schemes for branches and tips in the tree on the left is the same than for the gene lists on the right. Individuals with 2ISPs combining more than one type (polymorphic individuals) are black-rimmed; interestingly, their 2ISP patterns seem to cover always only two principal genotypes. Individuals where the 2ISPs cover apparent ancestor-descendant pairs (gradual shift) are indicated by colour gradients. E.g. the Ehime crenata shows a P4 genotype only found in this species (full orange circle), while the other two individuals show a transition from the ancestral subgenus Fagus genotype into the crenata-specific one (grey-orange gradient). The lucida P4 genotypes are fairly ancestral (mostly grey, with a brown strife) except for the Zhejiang individual, which shows the longipetiolata-specific genotype (blue).
It's obvious that at least some of the genes have disparate histories: their basic divergence patterns simply don't fit the combined tree, some are even in strong conflict.
Take gene P14, for instance. Both subgenera have their highly characteristic gene variants with relatively little variation within each variant. In subgenus Engleriana, the ancestral variant (pink) is gradually replaced by more specific ones; but only in F. engleriana (violet) and F. japonica (purple). Despite its tiny population size and isolation facilitating genetic drift, F. multinervis just kept the ancestral genotype of the subgenus. Within subgenus Fagus, the deep split between Eurasia (yellow) and North America (red) is well represented. With two notable exceptions: two F. mexicana have the Eurasian type (yellow; with a few specific modifications: dark orange), and F. sylvatica (s.str.) shows a distinct sequence type (red-green), which can be directly derived from the North American variant (full red). It's closeby sister species, the Western orientalis, however has the same (underived) genotype than all other Eurasian Fagus species (yellow). All data, genes, morphology, and fossil record, corroborate an old split between the Eurasian and North American species of subgenus Fagus (Denk & Grimm, 2009; Renner et al. 2016; Jiang et al., 2021, although 20 myrs off), hence, secondary contact and gene flow between the grandifolia-mexicana lineage and (part) of the sylvatica-orientalis lineage is the only explanation for what we see in gene P14. Convergent evolution can be ruled out, it's too many too consistent mutations for a gene with very low divergence to start with (no resolution within the main clades).
This particular secondary contact can even be pinpointed. It most likely occurred in the late Miocene. Beeches did cross the North Atlantic land bridge until into the Neogene. Palaeobotanists can even name the fossil, F. gussonii, physically linking Iceland with south-western Europe in the Miocene (Grímsson & Denk, 2005; Denk & Grimm, 2009). Fagus gussonii doesn't fit within the morphological space of the modern and ancient western Eurasian species but could well be a (morphologically primitive) member of the North American beech lineage (that includes modern-day grandifolia-mexicana). And it was coeval and sympatric with F. haidingeri, the precursor of all western Eurasian species that evolved from F. castaneifolia, the species that migrated from East Asia into western Eurasia in the Oligocene.
Fagus gussonii may have been the taxonomic vector that, after undergoing a couple of bottlenecks having crossed thousands of kilometers, hence the distinct satellite genotype, brought some North American gene variants into the European gene pool, before the North American lineage died out in Europe.
Vice versa, the modern-day North American species descended from a (high-latitude) lineage that in the late Miocene was still thriving on both sides of the Beringian land bridge, and in contact with the Eurasian clade within subgenus Fagus. The western North American species of that lineage maybe extinct but their shared ancestral genes may have survived in F. mexicana.
P52 and F289 give similar patterns. To much of a coincidence to be ignored just because the combined tree unambiguously supports the Eurasian-North American split.
When many histories are forced in one
A common negligence in phylogenomic studies is to exclusively rely on multi-species coalescents when facing inter-gene incongruence. No matter how sophisticated, they share one assumption: there is a species (true) tree and inter-gene conflict is mainly a matter of incomplete lineage sorting and varying evolutionary rates. Which may work for many organisms, but not for extratropical tree genera, especially not wind-pollinated ones. One should be wary of reticulation. A tip: If you have the right data, use PhyloNet to reconstruct the multi-species coalescent network rather than to rely on *BEAST or ASTRAL (see also D. Morrison's GWoN post Getting the wrong tree when reticulations are ignored).
The now more and more often seen cloudograms reflect the topological ambiguity in the gene sample but not the amplitude of the inter-gene conflict. The rarely used super-networks (Supernetworks and gene incongruence) can show both.
Due to algorithmic limitations, this super-network is limited to four dimensions. Not all conflicting topological aspects found in the 28 gene trees are covered. It also includes an artefact, the light pink edge representing a splits between subgenus Engleriana + one F. longipetiolata individual and the remainder of subgenus Fagus. Nonetheless, this graph is much too boxy to be explained by incomplete lineage sorting, and much to spider-webby to fit with the fully resolved tree.
Exploratory data analysis using the gene map, the super-network and the BS consensus networks of the single-gene analysis gives us the much-needed hand to identify and unterstand the branching artefacts in the combined tree. Why did Jiang et al. – and I, using the de-constructed individiual-level sequence data – get e.g. an unambiguously supported East Asian clade, although the gene data produces only very faint character support for such clade? The whole data only includes three (semi-)conserved point mutations in three different genes. (Nonetheless, pushing a BS support of 57, 53 and 98 in those genes for an East Asian clade; and fail to resolve the species within the clade...)
The reason is: we force non-treelike data, slow-evolving, low-divergent genes with partly disparate histories and reflecting past gene flow, introgression and hybridisation, into a tree. Let's trace the decisions a tree inference has to make by starting with a star tree.
The first obvious branching of this tree is to seperate the two subgenera. Their members are most-distant in the majority of genes and each side differs from the other by a good number of rather conserved point mutations. Plus, there's a lot of overlapping patterns in the members of subgenus Engleriana. Any tree method, no matter how sophisticated (Neighbour-joining or multi-species coalescent), will go for this split.
By the way, any outgroup-defined root will connect at this split because any living relative of Fagus is a distant one (inevitable ingroup-outgroup attraction). The beech lineage split from the rest of the family at least 80 myrs ago (Grímsson et al. 2016). This root is not necessarily wrong but if it is, we have no chance to get it right.
Now that the subgenera are seperated, the inference has to sort their species. In subgenus Engleriana this is trivial: F. multinervis has the lowest amount of derived (uniquely-evolved) sequence patterns, it is sequentially closer to subgenus Fagus than any individual of the other two species. Which must then be sisters—a nice example of very local, tip-long-branch attraction. In the super-network, we can already see that this sister relationship is only one of many alternatives, and that especially F. engleriana is far from forming a genetically coherent species (it has also the highest ITS lineage I diversity). But F. multinervis and F. japonica have specific genetic features, the inference will segregate them, and as by-catch, all F. engleriana individuals form a clade.
Now comes the tricky bit, we enter the danger zone: subgenus Fagus. The first step is still easy. The best-supported, character-wise, split is between the Old (Eurasian Fagus) and New World (North American grandifolia-mexicana).
In analogy to F. multinervis on the other side of the aile, the much more pronounced primitive character of some gene variants in F. mexicana draws them towards their Eurasian siblings. The more evolved, genetically unique and more coherent F. grandifolia (s.str.) must be broken apart, with the result that our tree shows two reciprocally monophyletic species. A possible hypothesis. However, sequence-wise, F. grandifolia collects gradually evolving descendants, while F. mexicana conserved the original less sorted and more polymorphic ancestral gene pool of all North American beeches. Is F. mexicana the source of F. grandifolia s.str., i.e. paraphyletic? Possibly, or, F. mexicana was much more affected by gene flow from rather primitive, now extinct western North American beeches still close to the common ancestor(s) of subgenus Fagus (see palaeomaps below). Last possibility, F. mexicana is polymorph because it's a hybrid of the eastern North American F. grandifolia (s.str.) precursor and its extinct western North American relatives (the lineage that produced e.g. the Miocene F. idahoensis and F. washingtonensis, I/W in the palaeogeographic map above). To solve this, one needs much more divergent nuclear gene regions.
With the North American being placed, the western Eurasian species must be next.
Why? Remember: they actually share some unique and derived genotypes with the North Americans, so there is actual character support for a North America + western Eurasia | East Asia (spp. of both subgenera) split. The Engleriana are long firmly placed afar, which leaves the East Asian Fagus spp. The East Asian clade is nothing but the indirect product of the North Americans being very distinct but attracted to subgenus Engleriana, and the western Eurasians being distinct but too close to the North Americans because of (late) Miocene gene flow. But if we look at the actual gene sequence, we have no problem evolving the distinct western Eurasian gene variants directly from the ones still found in F. crenata. Why don't we get a split, or at least F. crenata as the next branch?
Because of the pecularities of the hayataes. They are, among the remaining species, the ones that have at least in common with the others. Well, only the insular ones, F. hayatae s.str. They don't fit in any subtree involving any of the other East Asian species. Hence, they tear their sister, F. pashanica, the continental ones with much more ambiguous genetic signatures – in some genes so primitive that all subgenus Fagus variants (North American, western Eurasian and East Asian) can be derived from them – away from the rest. Jiang et al.'s results, their trees, may "... not support the basal most position of F. hayatae within the genus Fagus" but a few of their genes conspiciously would fit such a scenario. There is an old stinch imprinted in the genome of F. pashanica.
Fagus crenata is the most unspecific of the remaining three, while individuals of F. longipetiolata and F. lucida are rich in unique sequence patterns inviting tip-long branch attraction. The latter two have nothing exclusive in common; there's not a single conserved basepair or consistent mutational pattern backing the unambiguously supported longipetiolata-lucida clade. It's a single of the 28 genes, F138, that produces a high BS (89) support for an exclusive longipetiolata-lucida clade. Looking at their gene variants, both seem to have independently evolved from the Eurasian Fagus ancestor; the main difference being that F. lucida was in closer contact with/remained closer to F. crenata. The combination of local LBA, unique mutations found in most or all individuals of each species, and secondary (!) gene exchange, i.e. hybridisation and introgression transferring genetic characteristics from one species into one or two individuals of the other, leaves no escape for the tree inference than to conclude that these two must be sisters. Which automatically determines the placement of F. crenata, the last tip.
And here we go: a fully resolved, fine-supported tree, inference-wise trivial tree
- in which only very few clades are monophyletic in the original sense, i.e. share an inclusive common origin (see also D. Morrison's Let's distinguish between Hennig and Cladistics; and Clades, cladograms, cladistics, and why networks are inevitable);
- with the one or other a data/branching artefact: false positives;
- and not showing any bit of the compelling evidence for reticulate evolution that shaped the modern species.
A preliminary species network of beeches
But if we explore the data, and add it to evidence from earlier single-gene data, we can put all the pieces together that build up the puzzling nuclear-genetic mosaics that are modern-day beeches.
| Only a doodle (technically a coral network metaphor), summarising the genetic differentation patterns in the old, cloned ITS data (Denk et al., 2002, 2005; Grimm & Denk, 2007), more recently assembled LEAFY intron (Oh et al., 2016; Renner et al., 2016; Worth et al., 2021) and 5S-IGS data (Cardoni et al., 2021), and the (unphased) 28-gene data of Jiang et al. (2021; Grimm, 2021) |
It's not a species tree, of course. But a species network. What did Jiang et al. (2021) write again in their discussion section 4.2?
In spite of these [evidence for reticulation among western Eurasian beech spp.], contemporary hybridization events in the genus should be considered the exception and not the norm, both within and outside China.
Contemporary maybe (but how does one explain the poor nuclear sorting of e.g. F. engleriana?) In general, it couldn't be farther from the reality of their own data.
Especially when it comes to the formation and establishment of the modern-day Chinese species.
The only fossil with affinities to F. lucida comes from Central Asia, which may explain the lucida-unique features but also its affinity to F. crenata, the eastern child of the once widespread F. castaneifolia (CAS/orange field in the palaeogeographic map below).
Fagus crenata is the only dominant, and most abundant East Asian beech species, and morphologically most similar to the western Eurasian beeches. Not Chinese today, their common ancestor spanned across Eurasia. A large active population size and wide range over long time periods explains the lack of specific genetic sequence features and seemingly random links to any other Eurasian species including all Chinese ones (and the picking up of at least five different plastomes on the way; Worth et al., 2021).
Fagus longipetiolata has an old morphology, essentially unchanged since the Oligocene-Miocene, and shows a striking combination of very specific and shared with F. crenata or picked up from F. lucida sequence features.
And finally, the politically impossible – for authors funded by the PRC government – but clear genetic split between the continental and insular populations of F. hayatae—a case of cryptic ongoing speciation (see also Grimm & Denk, 2014, for a case of cryptic speciation in maples). Morphologically indistinguishable, their morphology sets them apart from both F. lucida and F. longipetiolata. And there are hayatae-wide shared and unique genetic features. To maintain their morphological distinctness and genetic particularity, Fagus hayatae (s.str.) and F. pashanica must have had a (relatively inclusive) common origin; they are monophyletic in the best-possible sense. But their nucleomes tell two amazingly different stories about what happened after the species established. The continental part seems to be relict of much larger population, still carrying around more primitive gene variants: they have been closer to the source and may have exchanged genetic material with nearby, ±ancient species (hence the ITS dimorphism shared with F. longipetiolata). The insular ones must have gotten isolated relatively early and because of their always much smaller-over-time population size, they underwent increased genetic drift. They best exemplify how to "accumulate private mutations" as Jiang et al. point out for the "segregates recognized" (i.e. those not clashing with the Party's One-China politics).
Even though the analyses and new conclusions in the Jiang et al.'s paper are largely for the bin (they not only included a dating using too young constraints but also a biogeographic analysis with most surprising results), the produced data itself is just gorgeous. One only has to give it a careful, proper, open-minded look rather than to dump it mindlessly in any available inference black-box. The latter has, admittedly, a long tradition in phylogenetics and is becoming a self-runner in the age of Big Data.
Data links
The data matrices, trees and bootstrap consensus networks can be found in my figshare Fagaceae collection (version 3, updated 29/9/2021):
Grimm G. 2020. Fagaceae collection. figshare. Dataset. https://doi.org/10.6084/m9.figshare.11603547.v3
The tabulation (XLSX multli-spread-sheet file) of sequence patterns, data-/ run-statistics etc. will be included as Supplement file S5 in:
Cardoni S, Piredda R, Denk T, Grimm G, Papageorgiou AC, Schulze E-D, Scoppola A, Shanjani PS, Suyama Y, Tomaru N, Worth JRP, Simeone MC. 2021. Data for Cardoni et al. (2021): High-Throughput Sequencing of 5S-IGS rDNA in Fagus L. figshare. Dataset. https://doi.org/10.6084/m9.figshare.13793744.v2References
Cardoni S, Piredda R, Denk T, Grimm GW, Papageorgiou AC, Schulze E-D, Scoppola A, Shanjani PS, Suyama Y, Tomaru N, Worth JRP, Simeone MC. 2021. High-Throughput Sequencing of 5S-IGS rDNA in Fagus L. (Fagaceae) reveals complex evolutionary patterns and hybrid origin of modern species. bioRxiv, doi: 10.1101/2021.1102.1126.433057.
Denk T, Grimm G, Stögerer K, Langer M, Hemleben V. 2002. The evolutionary history of Fagus in western Eurasia: Evidence from genes, morphology and the fossil record. Plant Systematics and Evolution 232:213-236.
Denk T, Grimm GW, Hemleben V. 2005. Patterns of molecular and morphological differentiation in Fagus: implications for phylogeny. American Journal of Botany 92:1006–1016.
Grimm GW, Denk T. 2014. The Colchic region as refuge for relict tree lineages: cryptic speciation in field maples. Turkish Journal of Botany 38:1050–1066. [PDF]
Grimm GW, Denk T, Hemleben V. 2007. Coding of intraspecific nucleotide polymorphisms: a tool to resolve reticulate evolutionary relationships in the ITS of beech trees (Fagus L., Fagaceae). Systematics and Biodiversity 5:291-309.
Grímsson F, Denk T. 2005. Fagus from the Miocene of Iceland: Systematics and biogeographical considerations. Review of Palaeobotany and Palynology 134:27-54.
Grímsson F, Denk T. 2007. Floristic turnover in Iceland from 15 to 6 Ma – extracting biogeographical signals from fossil floral assemblages. Journal of Biogeography 34:1490–1504.
Grímsson F, Grimm GW, Zetter R, Denk T. 2016. Cretaceous and Paleogene Fagaceae from North America and Greenland: evidence for a Late Cretaceous split between Fagus and the remaining Fagaceae. Acta Palaeobotanica 56:247–305.
Potts AJ, Hedderson TA, Grimm GW. 2014. Constructing phylogenies in the presence of intra-individual site polymorphisms (2ISPs) with a focus on the nuclear ribosomal cistron. Systematic Biology 63:1–16.
Renner SS, Grimm GW, Kapli P, Denk T. 2016. Species relationships and divergence times in beeches: New insights from the inclusion of 53 young and old fossils in a birth-death clock model. Philosophical Transactions of the Royal Society B 371:20150135.
Ribeiro T, Loureiro J, Santos C, Morais-Cecílio L. 2011. Evolution of rDNA FISH patterns in the Fagaceae. Tree Genetics and Genomes 7:1113–1122.
Worth, J. R. P., Ihara-Ujino, T., Grimm, G. W., Wei, F.-J., Simeone, M. C., Li, P., Marthick, J., Harrison, Peter A., …, Tomaru, N. (2021). Chloroplast genome sequencing reveals complex patterns of ancient and recent chloroplast sharing in Japanese Fagus. Presentation, http://dx.doi.org/10.13140/RG.2.2.10679.24485
















Just went through the whole post and I could tell how science is a rigged game. I am confused about what is considered to be integrity in science. as a new scientist I wonder what is going on.
ReplyDeleteAll non-profit sciences still have a generally high level of integrity. It's far from cloud-cuckoo land and there are very foul apples, but it's typically (getting fewer and fewer) old white males forcing their young researchers to follow their faulty ways. And there are of course self-interest groups, that purposely ride dead horses to keep the rubels rollings (German saying). I covered examples of this in my bad science category.
DeleteThe first major problem is the confidential peer review process. The Jiang et al. paper is not "bad science": the authors did, compared to many other papers in the field, a very good job—but only to an unwitting researcher's eye, who has no idea about the data and organism. We cannot tell how rigorous the review was, because there's no transparency in the review process. I didn't get this paper to review, neither my beech-knowing colleagues. Giving the spectacularity of the data, one may wonder why this paper has been published in Chinese journal with domestically inflated impact factor and not some fancier journal such as e.g. J Biogeogr. Maybe they tried, and got rejected. But JSE is a natural choice: even if there was a challenging review, the prominence of the last author would have assured, it can be largely ignored. Provincialism is a increasing problem in peer review, gradually replacing classic peer review imperalism, e.g. retired U.S.-based "experts" rejecting research just because of "poor English".
Second problem is fragmentation of science. How to find equally competent as unbiased reviewers? Being an editor of a (classic) journal is the worst job you can have.
The third major problem is publish-or-perish. I probably spend more time looking at the data than the many authors spend analysing it. Feeding data into a black box is a no-brainer and self-seller; understanding the result, finding the pitfalls, requires a lot of background and experience (not just "expertise"). Which the authors cannot have. Plus, the more complex your data, the more complex the paper, the less the chance you get it published. Editors prefer easy food for their readers, need impact, and impact comes with big names and simple, shiny stories. With beech, you open the evolutionary Pandora's Box and you go down the darkest fox-hole. Now, far the most phylogeneticists are tree-thinkers. It's trivial to read a tree and sell it to the audience. If the authors would have analysed their data properly, having very few templates to follow, they would have run into problems and possibly erred as well; there would have been very few positive-constructive reviewers available making such a paper publishable.
Which is, by the way, also the reason that I will hide away this analysis in a supplement to a paper and not waste time trying to publish a formal, confidentially peer-reviewed comment :) The only possibly competent reviewers of such a comment would be my (former) co-authors.